The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum
- PMID: 29069494
- PMCID: PMC5691383
- DOI: 10.1093/gigascience/gix097
The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum
Abstract
Common bread wheat, Triticum aestivum, has one of the most complex genomes known to science, with 6 copies of each chromosome, enormous numbers of near-identical sequences scattered throughout, and an overall haploid size of more than 15 billion bases. Multiple past attempts to assemble the genome have produced assemblies that were well short of the estimated genome size. Here we report the first near-complete assembly of T. aestivum, using deep sequencing coverage from a combination of short Illumina reads and very long Pacific Biosciences reads. The final assembly contains 15 344 693 583 bases and has a weighted average (N50) contig size of 232 659 bases. This represents by far the most complete and contiguous assembly of the wheat genome to date, providing a strong foundation for future genetic studies of this important food crop. We also report how we used the recently published genome of Aegilops tauschii, the diploid ancestor of the wheat D genome, to identify 4 179 762 575 bp of T. aestivum that correspond to its D genome components.
Keywords: PacBio sequencing; genome assembly; hybrid assembly; plant genomes; wheat genome.
© The Author 2017. Published by Oxford University Press.
Figures

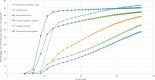
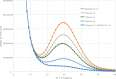
Similar articles
-
Independent assessment and improvement of wheat genome sequence assemblies using Fosill jumping libraries.Gigascience. 2018 May 1;7(5):giy053. doi: 10.1093/gigascience/giy053. Gigascience. 2018. PMID: 29762659 Free PMC article.
-
A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome.Science. 2014 Jul 18;345(6194):1251788. doi: 10.1126/science.1251788. Science. 2014. PMID: 25035500
-
An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations.Genome Res. 2017 May;27(5):885-896. doi: 10.1101/gr.217117.116. Genome Res. 2017. PMID: 28420692 Free PMC article.
-
[Progress on wheat A genome illustration and its evolutional analysis].Yi Chuan. 2019 Sep 20;41(9):836-844. doi: 10.16288/j.yczz.19-233. Yi Chuan. 2019. PMID: 31549682 Review. Chinese.
-
The Battle to Sequence the Bread Wheat Genome: A Tale of the Three Kingdoms.Genomics Proteomics Bioinformatics. 2020 Jun;18(3):221-229. doi: 10.1016/j.gpb.2019.09.005. Epub 2020 Jun 17. Genomics Proteomics Bioinformatics. 2020. PMID: 32561470 Free PMC article. Review.
Cited by
-
Fine mapping of the reduced height gene Rht22 in tetraploid wheat landrace Jianyangailanmai (Triticum turgidum L.).Theor Appl Genet. 2022 Oct;135(10):3643-3660. doi: 10.1007/s00122-022-04207-8. Epub 2022 Sep 4. Theor Appl Genet. 2022. PMID: 36057866
-
Genome-wide characterization and expression analysis of the CINNAMYL ALCOHOL DEHYDROGENASE gene family in Triticum aestivum.BMC Genomics. 2024 Aug 29;25(1):816. doi: 10.1186/s12864-024-10648-w. BMC Genomics. 2024. PMID: 39210247 Free PMC article.
-
Annotation of Siberian Larch (Larix sibirica Ledeb.) Nuclear Genome-One of the Most Cold-Resistant Tree Species in the Only Deciduous GENUS in Pinaceae.Plants (Basel). 2022 Aug 6;11(15):2062. doi: 10.3390/plants11152062. Plants (Basel). 2022. PMID: 35956540 Free PMC article.
-
Haplotype blocks for genomic prediction: a comparative evaluation in multiple crop datasets.Front Plant Sci. 2023 Sep 5;14:1217589. doi: 10.3389/fpls.2023.1217589. eCollection 2023. Front Plant Sci. 2023. PMID: 37731980 Free PMC article.
-
Identification of genes involved in male sterility in wheat (Triticum aestivum L.) which could be used in a genic hybrid breeding system.Plant Direct. 2020 Mar 10;4(3):e00201. doi: 10.1002/pld3.201. eCollection 2020 Mar. Plant Direct. 2020. PMID: 32181421 Free PMC article.
References
-
- International Wheat Genome Sequencing Consortium A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014;345(6194):1251788. - PubMed
-
- Li W, Zhang P, Fellers JP et al. . Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 2004;40(4):500–11. - PubMed
-
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep 1991;9(3):208–18.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

