Structural and Functional Insight into Canarypox Virus CNP058 Mediated Regulation of Apoptosis
- PMID: 29053589
- PMCID: PMC5691656
- DOI: 10.3390/v9100305
Structural and Functional Insight into Canarypox Virus CNP058 Mediated Regulation of Apoptosis
Abstract
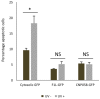
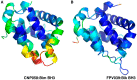
Programmed cell death or apoptosis is an important component of host defense systems against viral infection. The B-cell lymphoma 2 (Bcl-2) proteins family is the main arbiter of mitochondrially mediated apoptosis, and viruses have evolved sequence and structural mimics of Bcl-2 to subvert premature host cell apoptosis in response to viral infection. The sequencing of the canarypox virus genome identified a putative pro-survival Bcl-2 protein, CNP058. However, a role in apoptosis inhibition for CNP058 has not been identified to date. Here, we report that CNP058 is able to bind several host cell pro-death Bcl-2 proteins, including Bak and Bax, as well as several BH3 only-proteins including Bim, Bid, Bmf, Noxa, Puma, and Hrk with high to moderate affinities. We then defined the structural basis for CNP058 binding to pro-death Bcl-2 proteins by determining the crystal structure of CNP058 bound to Bim BH3. CNP058 adopts the conserved Bcl-2 like fold observed in cellular pro-survival Bcl-2 proteins, and utilizes the canonical ligand binding groove to bind Bim BH3. We then demonstrate that CNP058 is a potent inhibitor of ultraviolet (UV) induced apoptosis in a cell culture model. Our findings suggest that CNP058 is a potent inhibitor of apoptosis that is able to bind to BH3 domain peptides from a broad range of pro-death Bcl-2 proteins, and may play a key role in countering premature host apoptosis.
Keywords: Bcl-2; X-ray crystallography; apoptosis; avipoxvirus; isothermal titration calorimetry; poxvirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.J Biol Chem. 2018 Apr 13;293(15):5464-5477. doi: 10.1074/jbc.RA117.000591. Epub 2018 Feb 26. J Biol Chem. 2018. PMID: 29483196 Free PMC article.
-
Structural insight into tanapoxvirus-mediated inhibition of apoptosis.FEBS J. 2020 Sep;287(17):3733-3750. doi: 10.1111/febs.15365. Epub 2020 May 31. FEBS J. 2020. PMID: 32412687
-
Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk.Viruses. 2021 Jul 15;13(7):1374. doi: 10.3390/v13071374. Viruses. 2021. PMID: 34372579 Free PMC article.
-
Structural biology of the Bcl-2 family of proteins.Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
-
Structural biology of the Bcl-2 family and its mimicry by viral proteins.Cell Death Dis. 2013 Nov 7;4(11):e909. doi: 10.1038/cddis.2013.436. Cell Death Dis. 2013. PMID: 24201808 Free PMC article. Review.
Cited by
-
Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin.Viruses. 2019 Aug 27;11(9):789. doi: 10.3390/v11090789. Viruses. 2019. PMID: 31461953 Free PMC article.
-
Designing BH3-Mimetic Peptide Inhibitors for the Viral Bcl-2 Homologues A179L and BHRF1: Importance of Long-Range Electrostatic Interactions.ACS Omega. 2021 Oct 4;6(41):26976-26989. doi: 10.1021/acsomega.1c03385. eCollection 2021 Oct 19. ACS Omega. 2021. PMID: 34693118 Free PMC article.
-
Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.J Biol Chem. 2018 Apr 13;293(15):5464-5477. doi: 10.1074/jbc.RA117.000591. Epub 2018 Feb 26. J Biol Chem. 2018. PMID: 29483196 Free PMC article.
-
Poxviral Strategies to Overcome Host Cell Apoptosis.Pathogens. 2020 Dec 23;10(1):6. doi: 10.3390/pathogens10010006. Pathogens. 2020. PMID: 33374867 Free PMC article. Review.
-
The Bcl-2 Family: Ancient Origins, Conserved Structures, and Divergent Mechanisms.Biomolecules. 2020 Jan 12;10(1):128. doi: 10.3390/biom10010128. Biomolecules. 2020. PMID: 31940915 Free PMC article. Review.
References
-
- Kvansakul M., Hinds M.G. The structural biology of BH3-only proteins. Methods Enzymol. 2014;544:49–74. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

