Human TRPML1 channel structures in open and closed conformations
- PMID: 29019983
- PMCID: PMC5920536
- DOI: 10.1038/nature24036
Human TRPML1 channel structures in open and closed conformations
Abstract
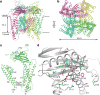
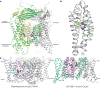
Transient receptor potential mucolipin 1 (TRPML1) is a Ca2+-releasing cation channel that mediates the calcium signalling and homeostasis of lysosomes. Mutations in TRPML1 lead to mucolipidosis type IV, a severe lysosomal storage disorder. Here we report two electron cryo-microscopy structures of full-length human TRPML1: a 3.72-Å apo structure at pH 7.0 in the closed state, and a 3.49-Å agonist-bound structure at pH 6.0 in an open state. Several aromatic and hydrophobic residues in pore helix 1, helices S5 and S6, and helix S6 of a neighbouring subunit, form a hydrophobic cavity to house the agonist, suggesting a distinct agonist-binding site from that found in TRPV1, a TRP channel from a different subfamily. The opening of TRPML1 is associated with distinct dilations of its lower gate together with a slight structural movement of pore helix 1. Our work reveals the regulatory mechanism of TRPML channels, facilitates better understanding of TRP channel activation, and provides insights into the molecular basis of mucolipidosis type IV pathogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





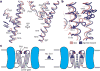
Similar articles
-
Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.Nature. 2017 Oct 19;550(7676):415-418. doi: 10.1038/nature24035. Epub 2017 Oct 11. Nature. 2017. PMID: 29019981 Free PMC article.
-
Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3.Nature. 2017 Oct 19;550(7676):411-414. doi: 10.1038/nature24055. Epub 2017 Oct 11. Nature. 2017. PMID: 29019979 Free PMC article.
-
Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism.Protein Cell. 2017 Nov;8(11):834-847. doi: 10.1007/s13238-017-0476-5. Epub 2017 Sep 21. Protein Cell. 2017. PMID: 28936784 Free PMC article.
-
The regulatory mechanism of mammalian TRPMLs revealed by cryo-EM.FEBS J. 2018 Jul;285(14):2579-2585. doi: 10.1111/febs.14443. Epub 2018 Apr 14. FEBS J. 2018. PMID: 29577631 Free PMC article. Review.
-
TRPML1: an ion channel in the lysosome.Handb Exp Pharmacol. 2014;222:631-45. doi: 10.1007/978-3-642-54215-2_24. Handb Exp Pharmacol. 2014. PMID: 24756723 Review.
Cited by
-
Ion channels as potential redox sensors in lysosomes.Channels (Austin). 2019 Dec;13(1):477-482. doi: 10.1080/19336950.2019.1684428. Channels (Austin). 2019. PMID: 31662029 Free PMC article. Review.
-
Lipid⁻Protein Interactions in Niemann⁻Pick Type C Disease: Insights from Molecular Modeling.Int J Mol Sci. 2019 Feb 7;20(3):717. doi: 10.3390/ijms20030717. Int J Mol Sci. 2019. PMID: 30736449 Free PMC article.
-
Lysosomes in senescence and aging.EMBO Rep. 2023 Nov 6;24(11):e57265. doi: 10.15252/embr.202357265. Epub 2023 Oct 9. EMBO Rep. 2023. PMID: 37811693 Free PMC article. Review.
-
Activation of the TRPML1 Ion Channel Induces Proton Secretion in the Human Gastric Parietal Cell Line HGT-1.Int J Mol Sci. 2024 Aug 13;25(16):8829. doi: 10.3390/ijms25168829. Int J Mol Sci. 2024. PMID: 39201515 Free PMC article.
-
MCOLN1/TRPML1 finely controls oncogenic autophagy in cancer by mediating zinc influx.Autophagy. 2021 Dec;17(12):4401-4422. doi: 10.1080/15548627.2021.1917132. Epub 2021 Apr 23. Autophagy. 2021. PMID: 33890549 Free PMC article.
References
-
- Zeevi DA, Frumkin A, Bach G. TRPML and lysosomal function. Biochim Biophys Acta. 2007;1772:851–858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

