Probing the interaction between NatA and the ribosome for co-translational protein acetylation
- PMID: 29016658
- PMCID: PMC5634638
- DOI: 10.1371/journal.pone.0186278
Probing the interaction between NatA and the ribosome for co-translational protein acetylation
Abstract
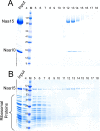
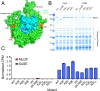
N-terminal acetylation is among the most abundant protein modifications in eukaryotic cells. Over the last decade, significant progress has been made in elucidating the function of N-terminal acetylation for a number of diverse systems, involved in a wide variety of biological processes. The enzymes responsible for the modification are the N-terminal acetyltransferases (NATs). The NATs are a highly conserved group of enzymes in eukaryotes, which are responsible for acetylating over 80% of the soluble proteome in human cells. Importantly, many of these NATs act co-translationally; they interact with the ribosome near the exit tunnel and acetylate the nascent protein chain as it is being translated. While the structures of many of the NATs have been determined, the molecular basis for the interaction with ribosome is not known. Here, using purified ribosomes and NatA, a very well-studied NAT, we show that NatA forms a stable complex with the ribosome in the absence of other stabilizing factors and through two conserved regions; primarily through an N-terminal domain and an internal basic helix. These regions may orient the active site of the NatA to face the peptide emerging from the exit tunnel. This work provides a framework for understanding how NatA and potentially other NATs interact with the ribosome for co-translational protein acetylation and sets the foundation for future studies to decouple N-terminal acetyltransferase activity from ribosome association.
Conflict of interest statement
Figures
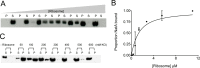




Similar articles
-
Protein N-Terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation.Trends Biochem Sci. 2021 Jan;46(1):15-27. doi: 10.1016/j.tibs.2020.08.005. Epub 2020 Sep 8. Trends Biochem Sci. 2021. PMID: 32912665 Free PMC article. Review.
-
Structure and Mechanism of Acetylation by the N-Terminal Dual Enzyme NatA/Naa50 Complex.Structure. 2019 Jul 2;27(7):1057-1070.e4. doi: 10.1016/j.str.2019.04.014. Epub 2019 May 30. Structure. 2019. PMID: 31155310 Free PMC article.
-
Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.Nat Struct Mol Biol. 2013 Sep;20(9):1098-105. doi: 10.1038/nsmb.2636. Epub 2013 Aug 4. Nat Struct Mol Biol. 2013. PMID: 23912279 Free PMC article.
-
A novel human NatA Nalpha-terminal acetyltransferase complex: hNaa16p-hNaa10p (hNat2-hArd1).BMC Biochem. 2009 May 29;10:15. doi: 10.1186/1471-2091-10-15. BMC Biochem. 2009. PMID: 19480662 Free PMC article.
-
Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases.Mol Cell. 2019 Mar 21;73(6):1097-1114. doi: 10.1016/j.molcel.2019.02.007. Epub 2019 Mar 13. Mol Cell. 2019. PMID: 30878283 Free PMC article. Review.
Cited by
-
Protein N-Terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation.Trends Biochem Sci. 2021 Jan;46(1):15-27. doi: 10.1016/j.tibs.2020.08.005. Epub 2020 Sep 8. Trends Biochem Sci. 2021. PMID: 32912665 Free PMC article. Review.
-
Structure of Human NatA and Its Regulation by the Huntingtin Interacting Protein HYPK.Structure. 2018 Jul 3;26(7):925-935.e8. doi: 10.1016/j.str.2018.04.003. Epub 2018 May 10. Structure. 2018. PMID: 29754825 Free PMC article.
-
NAA10 p.(D10G) and NAA10 p.(L11R) Variants Hamper Formation of the NatA N-Terminal Acetyltransferase Complex.Int J Mol Sci. 2020 Nov 26;21(23):8973. doi: 10.3390/ijms21238973. Int J Mol Sci. 2020. PMID: 33255974 Free PMC article.
-
Severe syndromic ID and skewed X-inactivation in a girl with NAA10 dysfunction and a novel heterozygous de novo NAA10 p.(His16Pro) variant - a case report.BMC Med Genet. 2020 Jul 22;21(1):153. doi: 10.1186/s12881-020-01091-1. BMC Med Genet. 2020. PMID: 32698785 Free PMC article.
-
Mechanisms of Congenital Heart Disease Caused by NAA15 Haploinsufficiency.Circ Res. 2021 Apr 16;128(8):1156-1169. doi: 10.1161/CIRCRESAHA.120.316966. Epub 2021 Feb 9. Circ Res. 2021. PMID: 33557580 Free PMC article. Clinical Trial.
References
-
- Aksnes H, Drazic A, Marie M, Arnesen T. First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem Sci. 2016;41(9):746–60. doi: 10.1016/j.tibs.2016.07.005 - DOI - PubMed
-
- Arnesen T, Gromyko D, Pendino F, Ryningen A, Varhaug JE, Lillehaug JR. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006;25(31):4350–60. Epub 2006/03/07. doi: 10.1038/sj.onc.1209469 . - DOI - PubMed
-
- Starheim KK, Gromyko D, Evjenth R, Ryningen A, Varhaug JE, Lillehaug JR, et al. Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol. 2009;29(13):3569–81. Epub 2009/04/29. doi: 10.1128/MCB.01909-08 ; PubMed Central PMCID: PMC2698767. - DOI - PMC - PubMed
-
- Pavlou D, Kirmizis A. Depletion of histone N-terminal-acetyltransferase Naa40 induces p53-independent apoptosis in colorectal cancer cells via the mitochondrial pathway. Apoptosis: an international journal on programmed cell death. 2016;21(3):298–311. doi: 10.1007/s10495-015-1207-0 - DOI - PMC - PubMed
-
- Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A, Kirmizis A. N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS Genet. 2013;9(9):e1003805 Epub 2013/09/27. doi: 10.1371/journal.pgen.1003805 ; PubMed Central PMCID: PMC3778019. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

