X-ray and EM structures of a natively glycosylated HIV-1 envelope trimer
- PMID: 28994411
- PMCID: PMC5633907
- DOI: 10.1107/S2059798317013353
X-ray and EM structures of a natively glycosylated HIV-1 envelope trimer
Abstract
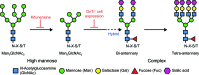
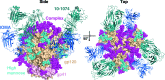
The structural and biochemical characterization of broadly neutralizing anti-HIV-1 antibodies (bNAbs) has been essential in guiding the design of potential vaccines to prevent infection by HIV-1. While these studies have revealed critical mechanisms by which bNAbs recognize and/or accommodate N-glycans on the trimeric envelope glycoprotein (Env), they have been limited to the visualization of high-mannose glycan forms only, since heterogeneity introduced from the presence of complex glycans makes it difficult to obtain high-resolution structures. 3.5 and 3.9 Å resolution crystal structures of the HIV-1 Env trimer with fully processed and native glycosylation were solved, revealing a glycan shield of high-mannose and complex-type N-glycans that were used to define the complete epitopes of two bNAbs. Here, the refinement of the N-glycans in the crystal structures is discussed and comparisons are made with glycan densities in glycosylated Env structures derived by single-particle cryo-electron microscopy.
Keywords: HIV-1 envelope; N-linked glycans; X-ray crystallography; single-particle cryo-EM.
Figures



Similar articles
-
Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site.Nat Struct Mol Biol. 2016 Oct;23(10):906-915. doi: 10.1038/nsmb.3291. Epub 2016 Sep 12. Nat Struct Mol Biol. 2016. PMID: 27617431 Free PMC article.
-
Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer.Acta Crystallogr D Biol Crystallogr. 2015 Oct;71(Pt 10):2099-108. doi: 10.1107/S1399004715013917. Epub 2015 Sep 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 26457433 Free PMC article.
-
Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope.Nat Commun. 2018 Mar 28;9(1):1251. doi: 10.1038/s41467-018-03632-y. Nat Commun. 2018. PMID: 29593217 Free PMC article.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
-
HIV-1 envelope glycoprotein structure.Curr Opin Struct Biol. 2013 Apr;23(2):268-76. doi: 10.1016/j.sbi.2013.03.007. Epub 2013 Apr 18. Curr Opin Struct Biol. 2013. PMID: 23602427 Free PMC article. Review.
Cited by
-
Towards Consistency in Geometry Restraints for Carbohydrates in the Pyranose form: Modern Dictionary Generators Reviewed.Curr Med Chem. 2022;29(7):1193-1207. doi: 10.2174/0929867328666210902140754. Curr Med Chem. 2022. PMID: 34477506 Free PMC article.
-
Online carbohydrate 3D structure validation with the Privateer web app.Acta Crystallogr F Struct Biol Commun. 2024 Feb 1;80(Pt 2):30-35. doi: 10.1107/S2053230X24000359. Epub 2024 Jan 24. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 38265073 Free PMC article.
-
Automatically Fixing Errors in Glycoprotein Structures with Rosetta.Structure. 2019 Jan 2;27(1):134-139.e3. doi: 10.1016/j.str.2018.09.006. Epub 2018 Oct 18. Structure. 2019. PMID: 30344107 Free PMC article.
-
Glycans in drug discovery.Medchemcomm. 2019 Jul 26;10(10):1678-1691. doi: 10.1039/c9md00292h. eCollection 2019 Oct 1. Medchemcomm. 2019. PMID: 31814952 Free PMC article. Review.
-
Subtle Longitudinal Alterations in Env Sequence Potentiate Differences in Sensitivity to Broadly Neutralizing Antibodies following Acute HIV-1 Subtype C Infection.J Virol. 2022 Dec 21;96(24):e0127022. doi: 10.1128/jvi.01270-22. Epub 2022 Dec 1. J Virol. 2022. PMID: 36453881 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

