Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1
- PMID: 28972166
- PMCID: PMC5704473
- DOI: 10.1074/jbc.M117.800706
Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1
Abstract
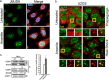
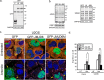
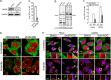
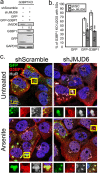
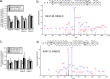
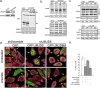
Stress granules (SG) are membrane-less organelles that are condensates of stalled translation initiation complexes and mRNAs. SG formation is a cytoprotective response to environmental stress and results from protein interactions involving regions of low amino acid complexity and poorly defined post-translational modifications of SG components. Many RNA-binding proteins are methylated, and we previously demonstrated that the potent SG-nucleating protein G3BP1 is methylated by protein arginine methyltransferase 1 and 5 (PRMT1 and PRMT5). G3BP1 methylation represses SG formation and is reversible. Here we functionally link JMJD6 (Jumonji C domain-containing protein 6) to G3BP1 demethylation. Our findings reveal that JMJD6 is a novel SG component that interacts with G3BP1 complexes, and its expression reduces G3BP1 monomethylation and asymmetric dimethylation at three Arg residues. Knockdown of JMJD6 repressed SG formation and G3BP1 demethylation, but SG formation and G3BP1 demethylation were rescued with catalytically active but not mutant JMJD6. These results suggest that JMJD6 functions directly or indirectly as an arginine demethylase of G3BP1 that promotes SG formation.
Keywords: RNA-binding protein; demethylase; post-translational modification (PTM); protein methylation; stress granule; stress response.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
Similar articles
-
The multi-functional RNA-binding protein G3BP1 and its potential implication in neurodegenerative disease.J Neurochem. 2021 May;157(4):944-962. doi: 10.1111/jnc.15280. Epub 2021 Jan 8. J Neurochem. 2021. PMID: 33349931 Free PMC article. Review.
-
Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly.J Biol Chem. 2016 Oct 21;291(43):22671-22685. doi: 10.1074/jbc.M116.739573. Epub 2016 Sep 6. J Biol Chem. 2016. PMID: 27601476 Free PMC article.
-
The Acetylation of Lysine-376 of G3BP1 Regulates RNA Binding and Stress Granule Dynamics.Mol Cell Biol. 2019 Oct 28;39(22):e00052-19. doi: 10.1128/MCB.00052-19. Print 2019 Nov 15. Mol Cell Biol. 2019. PMID: 31481451 Free PMC article.
-
UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1.Curr Biol. 2020 Feb 24;30(4):698-707.e6. doi: 10.1016/j.cub.2019.12.020. Epub 2020 Jan 16. Curr Biol. 2020. PMID: 31956030
-
The roles of G3BP1 in human diseases (review).Gene. 2022 May 5;821:146294. doi: 10.1016/j.gene.2022.146294. Epub 2022 Feb 14. Gene. 2022. PMID: 35176431 Review.
Cited by
-
Biomolecular condensates and disease pathogenesis.Sci China Life Sci. 2024 Sep;67(9):1792-1832. doi: 10.1007/s11427-024-2661-3. Epub 2024 Jul 17. Sci China Life Sci. 2024. PMID: 39037698 Review.
-
Ethylene -dependent and -independent superficial scald resistance mechanisms in 'Granny Smith' apple fruit.Sci Rep. 2018 Jul 30;8(1):11436. doi: 10.1038/s41598-018-29706-x. Sci Rep. 2018. PMID: 30061655 Free PMC article.
-
The multi-functional RNA-binding protein G3BP1 and its potential implication in neurodegenerative disease.J Neurochem. 2021 May;157(4):944-962. doi: 10.1111/jnc.15280. Epub 2021 Jan 8. J Neurochem. 2021. PMID: 33349931 Free PMC article. Review.
-
Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics.J Biol Chem. 2019 May 3;294(18):7137-7150. doi: 10.1074/jbc.TM118.001189. Epub 2018 Dec 26. J Biol Chem. 2019. PMID: 30587571 Free PMC article. Review.
-
The RGG motif proteins: Interactions, functions, and regulations.Wiley Interdiscip Rev RNA. 2023 Jan;14(1):e1748. doi: 10.1002/wrna.1748. Epub 2022 Jun 3. Wiley Interdiscip Rev RNA. 2023. PMID: 35661420 Free PMC article. Review.
References
-
- McDonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., and Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 - PubMed
-
- Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., and Harding H. P. (2003) Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284, C273–C284 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

