Coordinated repression of BIM and PUMA by Epstein-Barr virus latent genes maintains the survival of Burkitt lymphoma cells
- PMID: 28960205
- PMCID: PMC5762840
- DOI: 10.1038/cdd.2017.150
Coordinated repression of BIM and PUMA by Epstein-Barr virus latent genes maintains the survival of Burkitt lymphoma cells
Abstract
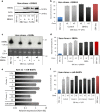
While the association of Epstein-Barr virus (EBV) with Burkitt lymphoma (BL) has long been recognised, the precise role of the virus in BL pathogenesis is not fully resolved. EBV can be lost spontaneously from some BL cell lines, and these EBV-loss lymphoma cells reportedly have a survival disadvantage. Here we have generated an extensive panel of EBV-loss clones from multiple BL backgrounds and examined their phenotype comparing them to their isogenic EBV-positive counterparts. We report that, while loss of EBV from BL cells is rare, it is consistently associated with an enhanced predisposition to undergo apoptosis and reduced tumorigenicity in vivo. Importantly, reinfection of EBV-loss clones with EBV, but surprisingly not transduction with individual BL-associated latent viral genes, restored protection from apoptosis. Expression profiling and functional analysis of apoptosis-related proteins and transcripts in BL cells revealed that EBV inhibits the upregulation of the proapoptotic BH3-only proteins, BIM and PUMA. We conclude that latent EBV genes cooperatively enhance the survival of BL cells by suppression of the intrinsic apoptosis pathway signalling via inhibition of the potent apoptosis initiators, BIM and PUMA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
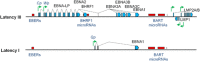
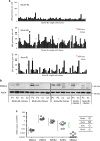
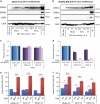
 ) and Mutu-BL (Δ) expressed relative to the endogenous control, GAPDH, and shown relative to the range seen in a panel of eight Latency I BL cell lines (grey bracket), including those from which the EBV-loss clones were isolated, as described elsewhere. EBNA2 expression is shown compared with the range seen in a panel of five LCLs (green bracket). EBNA1 refers to Q-U-K transcripts driven from the Qp promoter that are indicative of Latency I. BARTs refers to BamHI A transcripts that are spliced between exons 1 and 3 (the excised RNA gives rise to miR-BARTs)
) and Mutu-BL (Δ) expressed relative to the endogenous control, GAPDH, and shown relative to the range seen in a panel of eight Latency I BL cell lines (grey bracket), including those from which the EBV-loss clones were isolated, as described elsewhere. EBNA2 expression is shown compared with the range seen in a panel of five LCLs (green bracket). EBNA1 refers to Q-U-K transcripts driven from the Qp promoter that are indicative of Latency I. BARTs refers to BamHI A transcripts that are spliced between exons 1 and 3 (the excised RNA gives rise to miR-BARTs)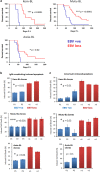
Similar articles
-
Combination of SAHA and bortezomib up-regulates CDKN2A and CDKN1A and induces apoptosis of Epstein-Barr virus-positive Wp-restricted Burkitt lymphoma and lymphoblastoid cell lines.Br J Haematol. 2014 Dec;167(5):639-50. doi: 10.1111/bjh.13089. Epub 2014 Aug 25. Br J Haematol. 2014. PMID: 25155625
-
Epstein-barr virus-induced resistance to drugs that activate the mitotic spindle assembly checkpoint in Burkitt's lymphoma cells.J Virol. 2007 Jan;81(1):248-60. doi: 10.1128/JVI.01096-06. Epub 2006 Oct 11. J Virol. 2007. PMID: 17035311 Free PMC article.
-
Epstein-Barr virus-encoded Bcl-2 homologue functions as a survival factor in Wp-restricted Burkitt lymphoma cell line P3HR-1.J Virol. 2010 Mar;84(6):2893-901. doi: 10.1128/JVI.01616-09. Epub 2009 Dec 30. J Virol. 2010. PMID: 20042495 Free PMC article.
-
Epstein-Barr virus, the TCL-1 oncogene and Burkitt's lymphoma.Trends Microbiol. 2003 Nov;11(11):495-7. doi: 10.1016/j.tim.2003.09.009. Trends Microbiol. 2003. PMID: 14607063 Review.
-
Role of Epstein-Barr virus in Burkitt's lymphoma.Curr Top Microbiol Immunol. 2001;258:141-51. doi: 10.1007/978-3-642-56515-1_9. Curr Top Microbiol Immunol. 2001. PMID: 11443859 Review.
Cited by
-
Infection-induced epigenetic changes and their impact on the pathogenesis of diseases.Semin Immunopathol. 2020 Apr;42(2):127-130. doi: 10.1007/s00281-020-00793-1. Semin Immunopathol. 2020. PMID: 32318841 Free PMC article. No abstract available.
-
Oncogenic Viruses and the Epigenome: How Viruses Hijack Epigenetic Mechanisms to Drive Cancer.Int J Mol Sci. 2023 May 31;24(11):9543. doi: 10.3390/ijms24119543. Int J Mol Sci. 2023. PMID: 37298494 Free PMC article. Review.
-
Burkitt Lymphomas Evolve to Escape Dependencies on Epstein-Barr Virus.Front Cell Infect Microbiol. 2021 Jan 11;10:606412. doi: 10.3389/fcimb.2020.606412. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33505922 Free PMC article. Review.
-
Emerging approaches to target mitochondrial apoptosis in cancer cells.F1000Res. 2019 Oct 24;8:F1000 Faculty Rev-1793. doi: 10.12688/f1000research.18872.1. eCollection 2019. F1000Res. 2019. PMID: 31681471 Free PMC article. Review.
-
The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms.Leukemia. 2022 Jul;36(7):1720-1748. doi: 10.1038/s41375-022-01620-2. Epub 2022 Jun 22. Leukemia. 2022. PMID: 35732829 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

