Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue
- PMID: 28950101
- PMCID: PMC5645043
- DOI: 10.1016/j.devcel.2017.08.024
Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue
Abstract
Mechanisms that control cell-cycle dynamics during tissue regeneration require elucidation. Here we find in zebrafish that regeneration of the epicardium, the mesothelial covering of the heart, is mediated by two phenotypically distinct epicardial cell subpopulations. These include a front of large, multinucleate leader cells, trailed by follower cells that divide to produce small, mononucleate daughters. By using live imaging of cell-cycle dynamics, we show that leader cells form by spatiotemporally regulated endoreplication, caused primarily by cytokinesis failure. Leader cells display greater velocities and mechanical tension within the epicardial tissue sheet, and experimentally induced tension anisotropy stimulates ectopic endoreplication. Unbalancing epicardial cell-cycle dynamics with chemical modulators indicated autonomous regenerative capacity in both leader and follower cells, with leaders displaying an enhanced capacity for surface coverage. Our findings provide evidence that mechanical tension can regulate cell-cycle dynamics in regenerating tissue, stratifying the source cell features to improve repair.
Keywords: endoreplication; epicardium; heart; mechanical tension; polyploidy; regeneration; zebrafish.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
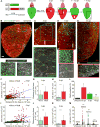
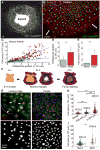

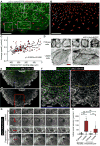


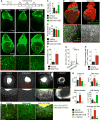
Comment in
-
Regeneration Tensed Up: Polyploidy Takes the Lead.Dev Cell. 2017 Sep 25;42(6):559-560. doi: 10.1016/j.devcel.2017.09.008. Dev Cell. 2017. PMID: 28950096
Similar articles
-
Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling.Nature. 2015 Jun 11;522(7555):226-230. doi: 10.1038/nature14325. Epub 2015 May 4. Nature. 2015. PMID: 25938716 Free PMC article.
-
Regeneration Tensed Up: Polyploidy Takes the Lead.Dev Cell. 2017 Sep 25;42(6):559-560. doi: 10.1016/j.devcel.2017.09.008. Dev Cell. 2017. PMID: 28950096
-
Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration.Development. 2016 Jan 15;143(2):232-43. doi: 10.1242/dev.130534. Epub 2015 Dec 10. Development. 2016. PMID: 26657776 Free PMC article.
-
Endoreplication: The Good, the Bad, and the Ugly.Trends Cell Biol. 2018 Jun;28(6):465-474. doi: 10.1016/j.tcb.2018.02.006. Epub 2018 Mar 19. Trends Cell Biol. 2018. PMID: 29567370 Free PMC article. Review.
-
The epicardium as a source of multipotent adult cardiac progenitor cells: Their origin, role and fate.Pharmacol Res. 2018 Jan;127:129-140. doi: 10.1016/j.phrs.2017.07.020. Epub 2017 Jul 24. Pharmacol Res. 2018. PMID: 28751220 Review.
Cited by
-
The recycling endosome protein Rab25 coordinates collective cell movements in the zebrafish surface epithelium.Elife. 2021 Mar 23;10:e66060. doi: 10.7554/eLife.66060. Elife. 2021. PMID: 33755014 Free PMC article.
-
Neutrophils facilitate the epicardial regenerative response after zebrafish heart injury.Dev Biol. 2024 Apr;508:93-106. doi: 10.1016/j.ydbio.2024.01.011. Epub 2024 Jan 28. Dev Biol. 2024. PMID: 38286185
-
Mechanics and regulation of cytokinetic abscission.Front Cell Dev Biol. 2022 Nov 24;10:1046617. doi: 10.3389/fcell.2022.1046617. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506096 Free PMC article. Review.
-
α-catenin mechanosensitivity as a route to cytokinesis failure through sequestration of LZTS2.bioRxiv [Preprint]. 2023 Aug 26:2023.08.25.554884. doi: 10.1101/2023.08.25.554884. bioRxiv. 2023. Update in: J Cell Biol. 2025 Mar 3;224(3):e202308124. doi: 10.1083/jcb.202308124. PMID: 37662204 Free PMC article. Updated. Preprint.
-
After wounding, a G-protein coupled receptor promotes the restoration of tension in epithelial cells.bioRxiv [Preprint]. 2024 Feb 18:2023.05.31.543122. doi: 10.1101/2023.05.31.543122. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 May 1;35(5):ar66. doi: 10.1091/mbc.E23-05-0204. PMID: 37398151 Free PMC article. Updated. Preprint.
References
-
- Abmayr SM, Balagopalan L, Galletta BJ, Hong SJ. Cell and molecular biology of myoblast fusion. Int. Rev. Cytol. 2003;225:33–89. - PubMed
-
- Akimoto S, Mitsumata M, Sasaguri T, Yoshida Y. Laminar shear stress inhibits vascular endothelial cell proliferation by inducing cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1) Circ. Res. 2000;86:185–90. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

