Customizable de novo design strategies for DOCK: Application to HIVgp41 and other therapeutic targets
- PMID: 28940386
- PMCID: PMC5659719
- DOI: 10.1002/jcc.25052
Customizable de novo design strategies for DOCK: Application to HIVgp41 and other therapeutic targets
Abstract
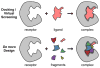
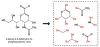
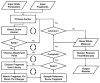
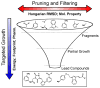
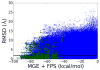
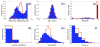
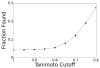
De novo design can be used to explore vast areas of chemical space in computational lead discovery. As a complement to virtual screening, from-scratch construction of molecules is not limited to compounds in pre-existing vendor catalogs. Here, we present an iterative fragment growth method, integrated into the program DOCK, in which new molecules are built using rules for allowable connections based on known molecules. The method leverages DOCK's advanced scoring and pruning approaches and users can define very specific criteria in terms of properties or features to customize growth toward a particular region of chemical space. The code was validated using three increasingly difficult classes of calculations: (1) Rebuilding known X-ray ligands taken from 663 complexes using only their component parts (focused libraries), (2) construction of new ligands in 57 drug target sites using a library derived from ∼13M drug-like compounds (generic libraries), and (3) application to a challenging protein-protein interface on the viral drug target HIVgp41. The computational testing confirms that the de novo DOCK routines are robust and working as envisioned, and the compelling results highlight the potential utility for designing new molecules against a wide variety of important protein targets. © 2017 Wiley Periodicals, Inc.
Keywords: DOCK; ZINC; chemical space; de novo design; drug discovery; footprint similarity; fragment libraries; scoring functions; structure-based design.
© 2017 Wiley Periodicals, Inc.
Figures


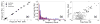
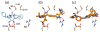
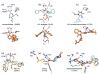


Similar articles
-
Strategies for lead discovery: application of footprint similarity targeting HIVgp41.Bioorg Med Chem. 2014 Jan 1;22(1):651-61. doi: 10.1016/j.bmc.2013.10.022. Epub 2013 Oct 26. Bioorg Med Chem. 2014. PMID: 24315195 Free PMC article.
-
Grid-based molecular footprint comparison method for docking and de novo design: application to HIVgp41.J Comput Chem. 2013 May 30;34(14):1226-1240. doi: 10.1002/jcc.23245. Epub 2013 Feb 22. J Comput Chem. 2013. PMID: 23436713 Free PMC article.
-
Implementation and evaluation of a docking-rescoring method using molecular footprint comparisons.J Comput Chem. 2011 Jul 30;32(10):2273-89. doi: 10.1002/jcc.21814. Epub 2011 May 3. J Comput Chem. 2011. PMID: 21541962 Free PMC article.
-
Docking and scoring in virtual screening for drug discovery: methods and applications.Nat Rev Drug Discov. 2004 Nov;3(11):935-49. doi: 10.1038/nrd1549. Nat Rev Drug Discov. 2004. PMID: 15520816 Review.
-
Chemistry-driven Hit-to-lead Optimization Guided by Structure-based Approaches.Mol Inform. 2018 Sep;37(9-10):e1800059. doi: 10.1002/minf.201800059. Epub 2018 Jul 27. Mol Inform. 2018. PMID: 30051601 Review.
Cited by
-
Virtual screening for small molecular non-covalent binders of the SARS-CoV-2 main protease.Arch Med Sci. 2021 Mar 19;17(3):838-842. doi: 10.5114/aoms/133122. eCollection 2021. Arch Med Sci. 2021. PMID: 34025857 Free PMC article. No abstract available.
-
Synthesis and Biological Evaluation of a Series of New Hybrid Amide Derivatives of Triazole and Thiazolidine-2,4-dione.Pharmaceuticals (Basel). 2024 Jun 3;17(6):723. doi: 10.3390/ph17060723. Pharmaceuticals (Basel). 2024. PMID: 38931390 Free PMC article.
-
DrugDevCovid19: An Atlas of Anti-COVID-19 Compounds Derived by Computer-Aided Drug Design.Molecules. 2022 Jan 21;27(3):683. doi: 10.3390/molecules27030683. Molecules. 2022. PMID: 35163948 Free PMC article. Review.
-
What Can De Novo Protein Design Bring to the Treatment of Hematological Disorders?Biology (Basel). 2023 Jan 20;12(2):166. doi: 10.3390/biology12020166. Biology (Basel). 2023. PMID: 36829445 Free PMC article. Review.
-
FastGrow: on-the-fly growing and its application to DYRK1A.J Comput Aided Mol Des. 2022 Sep;36(9):639-651. doi: 10.1007/s10822-022-00469-y. Epub 2022 Aug 22. J Comput Aided Mol Des. 2022. PMID: 35989379 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

