The eutheria-specific miR-290 cluster modulates placental growth and maternal-fetal transport
- PMID: 28935707
- PMCID: PMC5675445
- DOI: 10.1242/dev.151654
The eutheria-specific miR-290 cluster modulates placental growth and maternal-fetal transport
Abstract
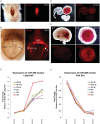
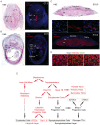
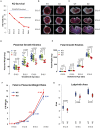
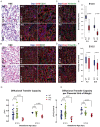
The vertebrate-specific ESCC microRNA family arises from two genetic loci in mammals: miR-290/miR-371 and miR-302. The miR-302 locus is found broadly among vertebrates, whereas the miR-290/miR-371 locus is unique to eutheria, suggesting a role in placental development. Here, we evaluate that role. A knock-in reporter for the mouse miR-290 cluster is expressed throughout the embryo until gastrulation, when it becomes specifically expressed in extraembryonic tissues and the germline. In the placenta, expression is limited to the trophoblast lineage, where it remains highly expressed until birth. Deletion of the miR-290 cluster gene (Mirc5) results in reduced trophoblast progenitor cell proliferation and a reduced DNA content in endoreduplicating trophoblast giant cells. The resulting placenta is reduced in size. In addition, the vascular labyrinth is disorganized, with thickening of the maternal-fetal blood barrier and an associated reduction in diffusion. Multiple mRNA targets of the miR-290 cluster microRNAs are upregulated. These data uncover a crucial function for the miR-290 cluster in the regulation of a network of genes required for placental development, suggesting a central role for these microRNAs in the evolution of placental mammals.
Keywords: Mouse; Placenta; Placentation; Trophoblast; miR-290; miR-371; microRNA.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

Similar articles
-
miR-126 regulates glycogen trophoblast proliferation and DNA methylation in the murine placenta.Dev Biol. 2019 May 1;449(1):21-34. doi: 10.1016/j.ydbio.2019.01.019. Epub 2019 Feb 14. Dev Biol. 2019. PMID: 30771304 Free PMC article.
-
Reduced Gene Dosage of Tfap2c Impairs Trophoblast Lineage Differentiation and Alters Maternal Blood Spaces in the Mouse Placenta.Biol Reprod. 2015 Aug;93(2):31. doi: 10.1095/biolreprod.114.126474. Epub 2015 Jun 10. Biol Reprod. 2015. PMID: 26063869
-
Trophoblast expression of fms-like tyrosine kinase 1 is not required for the establishment of the maternal-fetal interface in the mouse placenta.Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15637-42. doi: 10.1073/pnas.2635424100. Epub 2003 Dec 10. Proc Natl Acad Sci U S A. 2003. PMID: 14668430 Free PMC article.
-
The function of miR-519d in cell migration, invasion, and proliferation suggests a role in early placentation.Placenta. 2016 Dec;48:34-37. doi: 10.1016/j.placenta.2016.10.004. Epub 2016 Oct 10. Placenta. 2016. PMID: 27871470 Free PMC article. Review.
-
Review: The placenta and developmental programming: balancing fetal nutrient demands with maternal resource allocation.Placenta. 2012 Feb;33 Suppl:S23-7. doi: 10.1016/j.placenta.2011.11.013. Epub 2011 Dec 10. Placenta. 2012. PMID: 22154688 Review.
Cited by
-
Metazoan MicroRNAs.Cell. 2018 Mar 22;173(1):20-51. doi: 10.1016/j.cell.2018.03.006. Cell. 2018. PMID: 29570994 Free PMC article. Review.
-
The roles of microRNAs in mouse development.Nat Rev Genet. 2021 May;22(5):307-323. doi: 10.1038/s41576-020-00309-5. Epub 2021 Jan 15. Nat Rev Genet. 2021. PMID: 33452500 Review.
-
CPPED1-targeting microRNA-371a-5p expression in human placenta associates with spontaneous delivery.PLoS One. 2020 Jun 10;15(6):e0234403. doi: 10.1371/journal.pone.0234403. eCollection 2020. PLoS One. 2020. PMID: 32520951 Free PMC article.
-
MicroRNAs: key regulators of the trophoblast function in pregnancy disorders.J Assist Reprod Genet. 2023 Jan;40(1):3-17. doi: 10.1007/s10815-022-02677-9. Epub 2022 Dec 12. J Assist Reprod Genet. 2023. PMID: 36508034 Free PMC article. Review.
-
Placenta-derived macaque trophoblast stem cells: differentiation to syncytiotrophoblasts and extravillous trophoblasts reveals phenotypic reprogramming.Sci Rep. 2020 Nov 5;10(1):19159. doi: 10.1038/s41598-020-76313-w. Sci Rep. 2020. PMID: 33154556 Free PMC article.
References
-
- Biesterfeld S., Beckers S., Del Carmen Villa Cadenas M. and Schramm M. (2011). Feulgen staining remains the gold standard for precise DNA image cytometry. Anticancer Res. 31, 53-58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

