The Vibrio cholerae var regulon encodes a metallo-β-lactamase and an antibiotic efflux pump, which are regulated by VarR, a LysR-type transcription factor
- PMID: 28898293
- PMCID: PMC5595328
- DOI: 10.1371/journal.pone.0184255
The Vibrio cholerae var regulon encodes a metallo-β-lactamase and an antibiotic efflux pump, which are regulated by VarR, a LysR-type transcription factor
Abstract
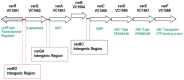
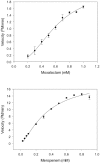
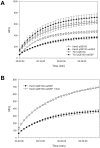
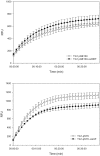
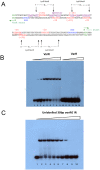
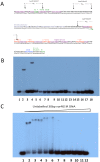
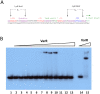
The genome sequence of V. cholerae O1 Biovar Eltor strain N16961 has revealed a putative antibiotic resistance (var) regulon that is predicted to encode a transcriptional activator (VarR), which is divergently transcribed relative to the putative resistance genes for both a metallo-β-lactamase (VarG) and an antibiotic efflux-pump (VarABCDEF). We sought to test whether these genes could confer antibiotic resistance and are organised as a regulon under the control of VarR. VarG was overexpressed and purified and shown to have β-lactamase activity against penicillins, cephalosporins and carbapenems, having the highest activity against meropenem. The expression of VarABCDEF in the Escherichia coli (ΔacrAB) strain KAM3 conferred resistance to a range of drugs, but most significant resistance was to the macrolide spiramycin. A gel-shift analysis was used to determine if VarR bound to the promoter regions of the resistance genes. Consistent with the regulation of these resistance genes, VarR binds to three distinct intergenic regions, varRG, varGA and varBC located upstream and adjacent to varG, varA and varC, respectively. VarR can act as a repressor at the varRG promoter region; whilst this repression was relieved upon addition of β-lactams, these did not dissociate the VarR/varRG-DNA complex, indicating that the de-repression of varR by β-lactams is indirect. Considering that the genomic arrangement of VarR-VarG is strikingly similar to that of AmpR-AmpC system, it is possible that V. cholerae has evolved a system for resistance to the newer β-lactams that would prove more beneficial to the bacterium in light of current selective pressures.
Conflict of interest statement
Figures
Similar articles
-
Transcriptional Repression of the VC2105 Protein by Vibrio FadR Suggests that It Is a New Auxiliary Member of the fad Regulon.Appl Environ Microbiol. 2016 Apr 18;82(9):2819-2832. doi: 10.1128/AEM.00293-16. Print 2016 May. Appl Environ Microbiol. 2016. PMID: 26944841 Free PMC article.
-
The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB.Gene. 1998 Mar 16;209(1-2):65-70. doi: 10.1016/s0378-1119(98)00018-3. Gene. 1998. PMID: 9524224
-
LysR family activator-regulated major facilitator superfamily transporters are involved in Vibrio cholerae antimicrobial compound resistance and intestinal colonisation.Int J Antimicrob Agents. 2013 Feb;41(2):188-92. doi: 10.1016/j.ijantimicag.2012.10.008. Epub 2012 Nov 30. Int J Antimicrob Agents. 2013. PMID: 23201336
-
Chromosomal beta-lactam resistance in enterobacteria.Scand J Infect Dis Suppl. 1986;49:38-45. Scand J Infect Dis Suppl. 1986. PMID: 3547624 Review.
-
Detection and analysis of gene expression during infection by in vivo expression technology.Philos Trans R Soc Lond B Biol Sci. 2000 May 29;355(1397):587-99. doi: 10.1098/rstb.2000.0600. Philos Trans R Soc Lond B Biol Sci. 2000. PMID: 10874732 Free PMC article. Review.
Cited by
-
Complete genome sequence and comparative analysis of a Vibrio vulnificus strain isolated from a clinical patient.Front Microbiol. 2023 Oct 31;14:1240835. doi: 10.3389/fmicb.2023.1240835. eCollection 2023. Front Microbiol. 2023. PMID: 38029170 Free PMC article.
-
Diversity and Proliferation of Metallo-β-Lactamases: a Clarion Call for Clinically Effective Metallo-β-Lactamase Inhibitors.Appl Environ Microbiol. 2018 Aug 31;84(18):e00698-18. doi: 10.1128/AEM.00698-18. Print 2018 Sep 15. Appl Environ Microbiol. 2018. PMID: 30006399 Free PMC article. Review.
-
Genomic insights into the 2022-2023Vibrio cholerae outbreak in Malawi.Nat Commun. 2024 Jul 26;15(1):6291. doi: 10.1038/s41467-024-50484-w. Nat Commun. 2024. PMID: 39060226 Free PMC article.
-
Characterization and Potentiating Effects of the Ethanolic Extracts of the Red Seaweed Gracillaria sp. on the Activity of Carbenicillin against Vibrios.ACS Omega. 2022 Dec 9;7(50):46486-46493. doi: 10.1021/acsomega.2c05288. eCollection 2022 Dec 20. ACS Omega. 2022. PMID: 36570316 Free PMC article.
-
Potential involvement of beta-lactamase homologous proteins in resistance to beta-lactam antibiotics in gram-negative bacteria of the ESKAPEE group.BMC Genomics. 2024 May 22;25(1):508. doi: 10.1186/s12864-024-10410-2. BMC Genomics. 2024. PMID: 38778284 Free PMC article.
References
-
- Ali M, Lopez AL, You YA, Kim YE, Sah B, Maskery B, et al. (2012) The global burden of cholera. Bulletin of the World Health Organization 90, 209–218A. doi: 10.2471/BLT.11.093427 - DOI - PMC - PubMed
-
- Childers BM, Klose KE (2007) Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future microbiology 2, 335–344. doi: 10.2217/17460913.2.3.335 - DOI - PubMed
-
- Odumosu O, Nicholas D, Yano H, Langridge W. (2010) AB toxins: a paradigm switch from deadly to desirable. Toxins 2, 1612–1645. doi: 10.3390/toxins2071612 - DOI - PMC - PubMed
-
- Sack DA, Lyke C, McLaughlin C, Suwanvanichkij V. (2001). Antimicrobial Resistance in Shigellosis, Cholera and Campylobacteriosis. Report No: WHO/CDS/CSR/DRS/2001/8. World Health Organization, Geneva.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

