Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling
- PMID: 28892079
- PMCID: PMC5790113
- DOI: 10.1038/ncb3610
Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling
Abstract
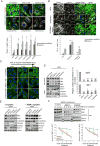
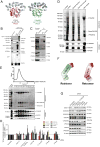
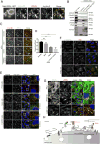
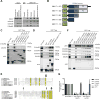
Following endocytosis into the endosomal network, integral membrane proteins undergo sorting for lysosomal degradation or are retrieved and recycled back to the cell surface. Here we describe the discovery of an ancient and conserved multiprotein complex that orchestrates cargo retrieval and recycling and, importantly, is biochemically and functionally distinct from the established retromer pathway. We have called this complex 'retriever'; it is a heterotrimer composed of DSCR3, C16orf62 and VPS29, and bears striking similarity to retromer. We establish that retriever associates with the cargo adaptor sorting nexin 17 (SNX17) and couples to CCC (CCDC93, CCDC22, COMMD) and WASH complexes to prevent lysosomal degradation and promote cell surface recycling of α5β1 integrin. Through quantitative proteomic analysis, we identify over 120 cell surface proteins, including numerous integrins, signalling receptors and solute transporters, that require SNX17-retriever to maintain their surface levels. Our identification of retriever establishes a major endosomal retrieval and recycling pathway.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Endosomal Trafficking: Retromer and Retriever Are Relatives in Recycling.Curr Biol. 2017 Nov 20;27(22):R1233-R1236. doi: 10.1016/j.cub.2017.10.004. Curr Biol. 2017. PMID: 29161566
Similar articles
-
Towards a molecular understanding of endosomal trafficking by Retromer and Retriever.Traffic. 2019 Jul;20(7):465-478. doi: 10.1111/tra.12649. Traffic. 2019. PMID: 30993794 Review.
-
Sorting nexin 17 (SNX17) links endosomal sorting to Eps15 homology domain protein 1 (EHD1)-mediated fission machinery.J Biol Chem. 2020 Mar 20;295(12):3837-3850. doi: 10.1074/jbc.RA119.011368. Epub 2020 Feb 10. J Biol Chem. 2020. PMID: 32041776 Free PMC article.
-
A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport.Nat Cell Biol. 2013 May;15(5):461-71. doi: 10.1038/ncb2721. Epub 2013 Apr 7. Nat Cell Biol. 2013. PMID: 23563491 Free PMC article.
-
Human Papillomavirus 16 L2 Recruits both Retromer and Retriever Complexes during Retrograde Trafficking of the Viral Genome to the Cell Nucleus.J Virol. 2021 Jan 13;95(3):e02068-20. doi: 10.1128/JVI.02068-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177206 Free PMC article.
-
Retromer and sorting nexins in endosomal sorting.Biochem Soc Trans. 2015 Feb;43(1):33-47. doi: 10.1042/BST20140290. Biochem Soc Trans. 2015. PMID: 25619244 Review.
Cited by
-
Cellular abundance of sodium phosphate cotransporter SLC20A1/PiT1 and phosphate uptake are controlled post-transcriptionally by ESCRT.J Biol Chem. 2022 Jun;298(6):101945. doi: 10.1016/j.jbc.2022.101945. Epub 2022 Apr 18. J Biol Chem. 2022. PMID: 35447110 Free PMC article.
-
Serotonin Receptor and Transporter Endocytosis Is an Important Factor in the Cellular Basis of Depression and Anxiety.Front Cell Neurosci. 2022 Feb 24;15:804592. doi: 10.3389/fncel.2021.804592. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35280519 Free PMC article.
-
TFEB controls retromer expression in response to nutrient availability.J Cell Biol. 2019 Dec 2;218(12):3954-3966. doi: 10.1083/jcb.201903006. Epub 2019 Nov 6. J Cell Biol. 2019. PMID: 31694921 Free PMC article.
-
Coronin2A links actin-based endosomal processes to the EHD1 fission machinery.Mol Biol Cell. 2022 Oct 1;33(12):ar107. doi: 10.1091/mbc.E21-12-0624. Epub 2022 Aug 3. Mol Biol Cell. 2022. PMID: 35921168 Free PMC article.
-
The endosomal system of primary human vascular endothelial cells and albumin-FcRn trafficking.J Cell Sci. 2023 Aug 1;136(15):jcs260912. doi: 10.1242/jcs.260912. Epub 2023 Aug 11. J Cell Sci. 2023. PMID: 37565427 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

