Cryo-EM Studies of Drp1 Reveal Cardiolipin Interactions that Activate the Helical Oligomer
- PMID: 28878368
- PMCID: PMC5587723
- DOI: 10.1038/s41598-017-11008-3
Cryo-EM Studies of Drp1 Reveal Cardiolipin Interactions that Activate the Helical Oligomer
Abstract
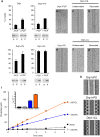
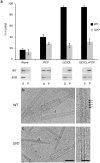
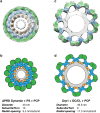
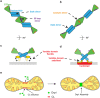
Dynamins are mechano-chemical GTPases involved in the remodeling of cellular membranes. In this study, we have investigated the mechanism of dynamin-related protein 1 (Drp1), a key mediator of mitochondrial fission. To date, it is unclear how Drp1 assembles on the mitochondrial outer membrane in response to different lipid signals to induce membrane fission. Here, we present cryo-EM structures of Drp1 helices on nanotubes with distinct lipid compositions to mimic membrane interactions with the fission machinery. These Drp1 polymers assemble exclusively through stalk and G-domain dimerizations, which generates an expanded helical symmetry when compared to other dynamins. Interestingly, we found the characteristic gap between Drp1 and the lipid bilayer was lost when the mitochondrial specific lipid cardiolipin was present, as Drp1 directly interacted with the membrane. Moreover, this interaction leads to a change in the helical structure, which alters G-domain interactions to enhance GTPase activity. These results demonstrate how lipid cues at the mitochondrial outer membrane (MOM) can alter Drp1 structure to activate the fission machinery.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Distinct Splice Variants of Dynamin-related Protein 1 Differentially Utilize Mitochondrial Fission Factor as an Effector of Cooperative GTPase Activity.J Biol Chem. 2016 Jan 1;291(1):493-507. doi: 10.1074/jbc.M115.680181. Epub 2015 Nov 17. J Biol Chem. 2016. PMID: 26578513 Free PMC article.
-
The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction.J Biol Chem. 2015 May 1;290(18):11692-703. doi: 10.1074/jbc.M114.610881. Epub 2015 Mar 13. J Biol Chem. 2015. PMID: 25770210 Free PMC article.
-
A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission.Mol Biol Cell. 2014 Jun 15;25(12):1905-15. doi: 10.1091/mbc.E14-02-0728. Epub 2014 Apr 30. Mol Biol Cell. 2014. PMID: 24790094 Free PMC article.
-
[Mechanism of mitochondrial fission - structure and function of Drp1 protein].Postepy Biochem. 2016;62(2):127-137. Postepy Biochem. 2016. PMID: 28132464 Review. Polish.
-
Splitting up the powerhouse: structural insights into the mechanism of mitochondrial fission.Cell Mol Life Sci. 2015 Oct;72(19):3695-707. doi: 10.1007/s00018-015-1950-y. Epub 2015 Jun 10. Cell Mol Life Sci. 2015. PMID: 26059473 Free PMC article. Review.
Cited by
-
Molecular mechanisms of mitochondrial dynamics.Nat Rev Mol Cell Biol. 2025 Feb;26(2):123-146. doi: 10.1038/s41580-024-00785-1. Epub 2024 Oct 17. Nat Rev Mol Cell Biol. 2025. PMID: 39420231 Review.
-
Disease-associated mutations in Drp1 have fundamentally different effects on the mitochondrial fission machinery.Hum Mol Genet. 2023 Jun 5;32(12):1975-1987. doi: 10.1093/hmg/ddad029. Hum Mol Genet. 2023. PMID: 36795043 Free PMC article.
-
The variable domain from dynamin-related protein 1 promotes liquid-liquid phase separation that enhances its interaction with cardiolipin-containing membranes.Protein Sci. 2023 Nov;32(11):e4787. doi: 10.1002/pro.4787. Protein Sci. 2023. PMID: 37743569 Free PMC article.
-
Dynamic properties of mitochondria during human corticogenesis.Development. 2021 Feb 19;148(4):dev194183. doi: 10.1242/dev.194183. Development. 2021. PMID: 33608250 Free PMC article. Review.
-
Mitochondrial dysfunctions in barth syndrome.IUBMB Life. 2019 Jul;71(7):791-801. doi: 10.1002/iub.2018. Epub 2019 Feb 11. IUBMB Life. 2019. PMID: 30746873 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

