Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
- PMID: 28827346
- PMCID: PMC5594662
- DOI: 10.1073/pnas.1705176114
Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Abstract
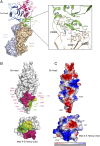
Severe fever with thrombocytopenia syndrome virus (SFTSV) and Rift Valley fever virus (RVFV) are two arthropod-borne phleboviruses in the Bunyaviridae family, which cause severe illness in humans and animals. Glycoprotein N (Gn) is one of the envelope proteins on the virus surface and is a major antigenic component. Despite its importance for virus entry and fusion, the molecular features of the phleboviruse Gn were unknown. Here, we present the crystal structures of the Gn head domain from both SFTSV and RVFV, which display a similar compact triangular shape overall, while the three subdomains (domains I, II, and III) making up the Gn head display different arrangements. Ten cysteines in the Gn stem region are conserved among phleboviruses, four of which are responsible for Gn dimerization, as revealed in this study, and they are highly conserved for all members in Bunyaviridae Therefore, we propose an anchoring mode on the viral surface. The complex structure of the SFTSV Gn head and human neutralizing antibody MAb 4-5 reveals that helices α6 in subdomain III is the key component for neutralization. Importantly, the structure indicates that domain III is an ideal region recognized by specific neutralizing antibodies, while domain II is probably recognized by broadly neutralizing antibodies. Collectively, Gn is a desirable vaccine target, and our data provide a molecular basis for the rational design of vaccines against the diseases caused by phleboviruses and a model for bunyavirus Gn embedding on the viral surface.
Keywords: RVFV; SFTSV; bunyavirus; glycoprotein; neutralizing antibody.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Similar articles
-
Characterization of Two Neutralizing Antibodies against Rift Valley Fever Virus Gn Protein.Viruses. 2020 Feb 27;12(3):259. doi: 10.3390/v12030259. Viruses. 2020. PMID: 32120864 Free PMC article.
-
The Role of Phlebovirus Glycoproteins in Viral Entry, Assembly and Release.Viruses. 2016 Jul 21;8(7):202. doi: 10.3390/v8070202. Viruses. 2016. PMID: 27455305 Free PMC article. Review.
-
An equine herpesvirus type 1 (EHV-1) vector expressing Rift Valley fever virus (RVFV) Gn and Gc induces neutralizing antibodies in sheep.Virol J. 2017 Aug 14;14(1):154. doi: 10.1186/s12985-017-0811-8. Virol J. 2017. PMID: 28807043 Free PMC article.
-
A Protective Monoclonal Antibody Targets a Site of Vulnerability on the Surface of Rift Valley Fever Virus.Cell Rep. 2018 Dec 26;25(13):3750-3758.e4. doi: 10.1016/j.celrep.2018.12.001. Cell Rep. 2018. PMID: 30590046 Free PMC article.
-
Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly.Viruses. 2014 Apr 21;6(4):1801-22. doi: 10.3390/v6041801. Viruses. 2014. PMID: 24755564 Free PMC article. Review.
Cited by
-
Antiviral activity and mechanism of the antifungal drug, anidulafungin, suggesting its potential to promote treatment of viral diseases.BMC Med. 2022 Oct 21;20(1):359. doi: 10.1186/s12916-022-02558-z. BMC Med. 2022. PMID: 36266654 Free PMC article.
-
Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.Nat Commun. 2023 Oct 10;14(1):6333. doi: 10.1038/s41467-023-41804-7. Nat Commun. 2023. PMID: 37816705 Free PMC article.
-
Characterization of Two Neutralizing Antibodies against Rift Valley Fever Virus Gn Protein.Viruses. 2020 Feb 27;12(3):259. doi: 10.3390/v12030259. Viruses. 2020. PMID: 32120864 Free PMC article.
-
Molecular insight into the neutralization mechanism of human-origin monoclonal antibody AH100 against Hantaan virus.J Virol. 2024 Aug 20;98(8):e0088324. doi: 10.1128/jvi.00883-24. Epub 2024 Jul 30. J Virol. 2024. PMID: 39078157 Free PMC article.
-
An Introduction to Rift Valley Fever Virus.Methods Mol Biol. 2024;2824:1-14. doi: 10.1007/978-1-0716-3926-9_1. Methods Mol Biol. 2024. PMID: 39039402
References
-
- Knipe DM, Howley PM. Fields Virology. 6th Ed. Vol 2 Wolters Kluwer/Lippincott Williams & Wilkins Health; Philadelphia: 2013.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

