Ultrasensitive mutation detection identifies rare residual cells causing acute myelogenous leukemia relapse
- PMID: 28825596
- PMCID: PMC5669556
- DOI: 10.1172/JCI91964
Ultrasensitive mutation detection identifies rare residual cells causing acute myelogenous leukemia relapse
Abstract
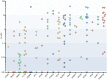
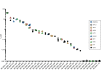
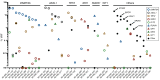
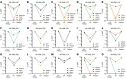
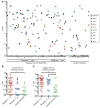
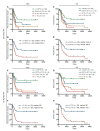
Acute myelogenous leukemia (AML) frequently relapses after complete remission (CR), necessitating improved detection and phenotypic characterization of treatment-resistant residual disease. In this work, we have optimized droplet digital PCR to broadly measure mutated alleles of recurrently mutated genes in CR marrows of AML patients at levels as low as 0.002% variant allele frequency. Most gene mutations persisted in CR, albeit at highly variable and gene-dependent levels. The majority of AML cases demonstrated residual aberrant oligoclonal hematopoiesis. Importantly, we detected very rare cells (as few as 1 in 15,000) that were genomically similar to the dominant blast populations at diagnosis and were fully clonally represented at relapse, identifying these rare cells as one common source of AML relapse. Clinically, the mutant allele burden was associated with overall survival in AML, and our findings narrow the repertoire of gene mutations useful in minimal residual disease-based prognostication in AML. Overall, this work delineates rare cell populations that cause AML relapse, with direct implications for AML research directions and strategies to improve AML therapies and outcome.
Conflict of interest statement
Figures
Similar articles
-
Molecular Minimal Residual Disease in Acute Myeloid Leukemia.N Engl J Med. 2018 Mar 29;378(13):1189-1199. doi: 10.1056/NEJMoa1716863. N Engl J Med. 2018. PMID: 29601269
-
Toward optimization of postremission therapy for residual disease-positive patients with acute myeloid leukemia.J Clin Oncol. 2008 Oct 20;26(30):4944-51. doi: 10.1200/JCO.2007.15.9814. Epub 2008 Jul 7. J Clin Oncol. 2008. PMID: 18606980
-
Core binding factor acute myeloid leukaemia and c-KIT mutations.Oncol Rep. 2013 May;29(5):1867-72. doi: 10.3892/or.2013.2328. Epub 2013 Mar 5. Oncol Rep. 2013. PMID: 23467883
-
Detection of minimal residual disease in acute myelogenous leukemia.Acta Haematol. 2004;112(1-2):40-54. doi: 10.1159/000077559. Acta Haematol. 2004. PMID: 15179004 Review.
-
Should the presence of minimal residual disease (MRD) in morphologic complete remission alter post-remission strategy in AML?Best Pract Res Clin Haematol. 2011 Dec;24(4):509-14. doi: 10.1016/j.beha.2011.09.006. Epub 2011 Nov 9. Best Pract Res Clin Haematol. 2011. PMID: 22127313 Review.
Cited by
-
Clinical significance of miR-372 and miR-495 in acute myeloid leukemia.Oncol Lett. 2020 Aug;20(2):1938-1944. doi: 10.3892/ol.2020.11748. Epub 2020 Jun 17. Oncol Lett. 2020. PMID: 32724438 Free PMC article.
-
Current and Emerging Techniques for Diagnosis and MRD Detection in AML: A Comprehensive Narrative Review.Cancers (Basel). 2023 Feb 21;15(5):1362. doi: 10.3390/cancers15051362. Cancers (Basel). 2023. PMID: 36900154 Free PMC article. Review.
-
Acute myeloid leukaemia.Lancet. 2018 Aug 18;392(10147):593-606. doi: 10.1016/S0140-6736(18)31041-9. Epub 2018 Aug 2. Lancet. 2018. PMID: 30078459 Free PMC article. Review.
-
DIP-microhaplotypes: new markers for detection of unbalanced DNA mixtures.Int J Legal Med. 2021 Jan;135(1):13-21. doi: 10.1007/s00414-020-02288-y. Epub 2020 May 5. Int J Legal Med. 2021. PMID: 32372232
-
The Prognostic Significance of Measurable ("Minimal") Residual Disease in Acute Myeloid Leukemia.Curr Hematol Malig Rep. 2017 Dec;12(6):547-556. doi: 10.1007/s11899-017-0420-z. Curr Hematol Malig Rep. 2017. PMID: 29027628 Review.
References
-
- Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood. 2004;104(10):3078–3085. doi: 10.1182/blood-2004-03-1036. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

