Mir505-3p regulates axonal development via inhibiting the autophagy pathway by targeting Atg12
- PMID: 28820282
- PMCID: PMC5640177
- DOI: 10.1080/15548627.2017.1353841
Mir505-3p regulates axonal development via inhibiting the autophagy pathway by targeting Atg12
Abstract
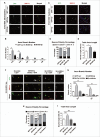
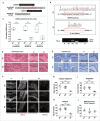
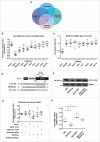
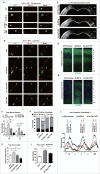
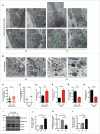
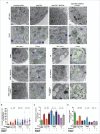
In addition to the canonical role in protein homeostasis, autophagy has recently been found to be involved in axonal dystrophy and neurodegeneration. Whether autophagy may also be involved in neural development remains largely unclear. Here we report that Mir505-3p is a crucial regulator for axonal elongation and branching in vitro and in vivo, through modulating autophagy in neurons. We identify that the key target gene of Mir505-3p in neurons is Atg12, encoding ATG12 (autophagy-related 12) which is an essential component of the autophagy machinery during the initiation and expansion steps of autophagosome formation. Importantly, axonal development is compromised in brains of mir505 knockout mice, in which autophagy signaling and formation of autophagosomes are consistently enhanced. These results define Mir505-3p-ATG12 as a vital signaling cascade for axonal development via the autophagy pathway, further suggesting the critical role of autophagy in neural development.
Keywords: ATG12; Mir505–3p; autophagy; axonal development; microRNA.
Figures


Similar articles
-
A SRSF1 self-binding mechanism restrains Mir505-3p from inhibiting proliferation of neural tumor cell lines.Anticancer Drugs. 2018 Jan;29(1):40-49. doi: 10.1097/CAD.0000000000000564. Anticancer Drugs. 2018. PMID: 29120871
-
Foot-and-mouth disease virus infection suppresses autophagy and NF-кB antiviral responses via degradation of ATG5-ATG12 by 3Cpro.Cell Death Dis. 2017 Jan 19;8(1):e2561. doi: 10.1038/cddis.2016.489. Cell Death Dis. 2017. PMID: 28102839 Free PMC article.
-
Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth.Cell Death Dis. 2017 Feb 9;8(2):e2605. doi: 10.1038/cddis.2017.31. Cell Death Dis. 2017. PMID: 28182000 Free PMC article.
-
miR-199a-3p is involved in estrogen-mediated autophagy through the IGF-1/mTOR pathway in osteocyte-like MLO-Y4 cells.J Cell Physiol. 2018 Mar;233(3):2292-2303. doi: 10.1002/jcp.26101. Epub 2017 Aug 25. J Cell Physiol. 2018. PMID: 28708244
-
Mechanisms of neuronal homeostasis: Autophagy in the axon.Brain Res. 2016 Oct 15;1649(Pt B):143-150. doi: 10.1016/j.brainres.2016.03.047. Epub 2016 Mar 30. Brain Res. 2016. PMID: 27038755 Free PMC article. Review.
Cited by
-
Effects of ambroxol on the autophagy-lysosome pathway and mitochondria in primary cortical neurons.Sci Rep. 2018 Jan 23;8(1):1385. doi: 10.1038/s41598-018-19479-8. Sci Rep. 2018. PMID: 29362387 Free PMC article.
-
Molecular Mechanism and Regulation of Autophagy and Its Potential Role in Epilepsy.Cells. 2022 Aug 23;11(17):2621. doi: 10.3390/cells11172621. Cells. 2022. PMID: 36078029 Free PMC article. Review.
-
MicroRNA in Brain pathology: Neurodegeneration the Other Side of the Brain Cancer.Noncoding RNA. 2019 Feb 23;5(1):20. doi: 10.3390/ncrna5010020. Noncoding RNA. 2019. PMID: 30813461 Free PMC article. Review.
-
Autophagy in axonal and presynaptic development.Curr Opin Neurobiol. 2021 Aug;69:139-148. doi: 10.1016/j.conb.2021.03.011. Epub 2021 Apr 30. Curr Opin Neurobiol. 2021. PMID: 33940492 Free PMC article. Review.
-
Analyses of single-cell and bulk RNA sequencing combined with machine learning reveal the expression patterns of disrupted mitophagy in schizophrenia.Front Psychiatry. 2024 Sep 17;15:1429437. doi: 10.3389/fpsyt.2024.1429437. eCollection 2024. Front Psychiatry. 2024. PMID: 39355378 Free PMC article.
References
-
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. https://doi.org/10.1016/j.cell.2007.12.018. PMID:18191218 - DOI - PMC - PubMed
-
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4, pii: a009886. https://doi.org/10.1101/cshperspect.a009886. PMID:22258914. - DOI - PMC - PubMed
-
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al.. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880-4. https://doi.org/10.1038/nature04723. PMID:16625205 - DOI - PubMed
-
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al.. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885-9. https://doi.org/10.1038/nature04724. PMID:16625204 - DOI - PubMed
-
- Komatsu M, Waguri S, Koike M, Sou Y, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al.. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149-63. https://doi.org/10.1016/j.cell.2007.10.035. PMID:18083104 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
