EB1 and EB3 regulate microtubule minus end organization and Golgi morphology
- PMID: 28814570
- PMCID: PMC5626540
- DOI: 10.1083/jcb.201701024
EB1 and EB3 regulate microtubule minus end organization and Golgi morphology
Abstract
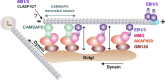
End-binding proteins (EBs) are the core components of microtubule plus end tracking protein complexes, but it is currently unknown whether they are essential for mammalian microtubule organization. Here, by using CRISPR/Cas9-mediated knockout technology, we generated stable cell lines lacking EB2 and EB3 and the C-terminal partner-binding half of EB1. These cell lines show only mild defects in cell division and microtubule polymerization. However, the length of CAMSAP2-decorated stretches at noncentrosomal microtubule minus ends in these cells is reduced, microtubules are detached from Golgi membranes, and the Golgi complex is more compact. Coorganization of microtubules and Golgi membranes depends on the EB1/EB3-myomegalin complex, which acts as membrane-microtubule tether and counteracts tight clustering of individual Golgi stacks. Disruption of EB1 and EB3 also perturbs cell migration, polarity, and the distribution of focal adhesions. EB1 and EB3 thus affect multiple interphase processes and have a major impact on microtubule minus end organization.
© 2017 Yang et al.
Figures

Similar articles
-
Molecular Pathway of Microtubule Organization at the Golgi Apparatus.Dev Cell. 2016 Oct 10;39(1):44-60. doi: 10.1016/j.devcel.2016.08.009. Epub 2016 Sep 22. Dev Cell. 2016. PMID: 27666745
-
Noncentrosomal microtubules regulate autophagosome transport through CAMSAP2-EB1 cross-talk.FEBS Lett. 2017 Aug;591(16):2379-2393. doi: 10.1002/1873-3468.12758. Epub 2017 Aug 9. FEBS Lett. 2017. PMID: 28726242
-
A newly identified myomegalin isoform functions in Golgi microtubule organization and ER-Golgi transport.J Cell Sci. 2014 Nov 15;127(Pt 22):4904-17. doi: 10.1242/jcs.155408. Epub 2014 Sep 12. J Cell Sci. 2014. PMID: 25217626 Free PMC article.
-
Microtubule plus ends, motors, and traffic of Golgi membranes.Biochim Biophys Acta. 2005 Jul 10;1744(3):316-24. doi: 10.1016/j.bbamcr.2005.05.001. Biochim Biophys Acta. 2005. PMID: 15950296 Review.
-
Regulation of end-binding protein EB1 in the control of microtubule dynamics.Cell Mol Life Sci. 2017 Jul;74(13):2381-2393. doi: 10.1007/s00018-017-2476-2. Epub 2017 Feb 15. Cell Mol Life Sci. 2017. PMID: 28204846 Free PMC article. Review.
Cited by
-
Identification of Functional Interactome of Gastric Cancer Cells with Helicobacter pylori Outer Membrane Protein HpaA by HPLC-MS/MS.Biomed Res Int. 2020 Jun 5;2020:1052926. doi: 10.1155/2020/1052926. eCollection 2020. Biomed Res Int. 2020. PMID: 32566649 Free PMC article.
-
CG-NAP/Kinase Interactions Fine-Tune T Cell Functions.Front Immunol. 2019 Nov 12;10:2642. doi: 10.3389/fimmu.2019.02642. eCollection 2019. Front Immunol. 2019. PMID: 31781123 Free PMC article. Review.
-
p73 regulates ependymal planar cell polarity by modulating actin and microtubule cytoskeleton.Cell Death Dis. 2018 Dec 5;9(12):1183. doi: 10.1038/s41419-018-1205-6. Cell Death Dis. 2018. PMID: 30518789 Free PMC article.
-
Optogenetic EB1 inactivation shortens metaphase spindles by disrupting cortical force-producing interactions with astral microtubules.Curr Biol. 2022 Mar 14;32(5):1197-1205.e4. doi: 10.1016/j.cub.2022.01.017. Epub 2022 Jan 31. Curr Biol. 2022. PMID: 35090591 Free PMC article.
-
Intraflagellar transport 20 promotes collective cancer cell invasion by regulating polarized organization of Golgi-associated microtubules.Cancer Sci. 2019 Apr;110(4):1306-1316. doi: 10.1111/cas.13970. Epub 2019 Feb 28. Cancer Sci. 2019. PMID: 30742741 Free PMC article.
References
-
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., and Galjart N.. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935. 10.1016/S0092-8674(01)00288-4 - DOI - PubMed
-
- Ban R., Matsuzaki H., Akashi T., Sakashita G., Taniguchi H., Park S.Y., Tanaka H., Furukawa K., and Urano T.. 2009. Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J. Biol. Chem. 284:28367–28381. 10.1074/jbc.M109.000273 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

