Molecular Rationale behind the Differential Substrate Specificity of Bacterial RND Multi-Drug Transporters
- PMID: 28808284
- PMCID: PMC5556075
- DOI: 10.1038/s41598-017-08747-8
Molecular Rationale behind the Differential Substrate Specificity of Bacterial RND Multi-Drug Transporters
Abstract
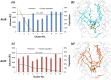
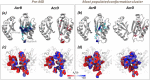
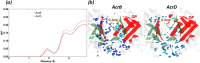
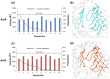
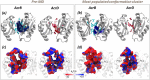
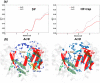
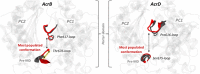
Resistance-Nodulation-cell Division (RND) transporters AcrB and AcrD of Escherichia coli expel a wide range of substrates out of the cell in conjunction with AcrA and TolC, contributing to the onset of bacterial multidrug resistance. Despite sharing an overall sequence identity of ~66% (similarity ~80%), these RND transporters feature distinct substrate specificity patterns whose underlying basis remains elusive. We performed exhaustive comparative analyses of the putative substrate binding pockets considering crystal structures, homology models and conformations extracted from multi-copy μs-long molecular dynamics simulations of both AcrB and AcrD. The impact of physicochemical and topographical properties (volume, shape, lipophilicity, electrostatic potential, hydration and distribution of multi-functional sites) within the pockets on their substrate specificities was quantitatively assessed. Differences in the lipophilic and electrostatic potentials among the pockets were identified. In particular, the deep pocket of AcrB showed the largest lipophilicity convincingly pointing out its possible role as a lipophilicity-based selectivity filter. Furthermore, we identified dynamic features (not inferable from sequence analysis or static structures) such as different flexibilities of specific protein loops that could potentially influence the substrate recognition and transport profile. Our findings can be valuable for drawing structure (dynamics)-activity relationship to be employed in drug design.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

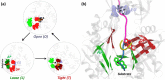

Similar articles
-
Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops.J Bacteriol. 2002 Dec;184(23):6490-8. doi: 10.1128/JB.184.23.6490-6499.2002. J Bacteriol. 2002. PMID: 12426336 Free PMC article.
-
Substrate-dependent dynamics of the multidrug efflux transporter AcrB of Escherichia coli.Sci Rep. 2016 Feb 26;6:21909. doi: 10.1038/srep21909. Sci Rep. 2016. PMID: 26916090 Free PMC article.
-
Computer simulations suggest direct and stable tip to tip interaction between the outer membrane channel TolC and the isolated docking domain of the multidrug RND efflux transporter AcrB.Biochim Biophys Acta. 2016 Jul;1858(7 Pt A):1419-26. doi: 10.1016/j.bbamem.2016.03.029. Epub 2016 Apr 2. Biochim Biophys Acta. 2016. PMID: 27045078
-
The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance.Curr Drug Targets. 2008 Sep;9(9):729-49. doi: 10.2174/138945008785747789. Curr Drug Targets. 2008. PMID: 18781920 Review.
-
RND efflux pumps: structural information translated into function and inhibition mechanisms.Curr Top Med Chem. 2013;13(24):3079-100. doi: 10.2174/15680266113136660220. Curr Top Med Chem. 2013. PMID: 24200360 Review.
Cited by
-
Chlorpromazine and Amitriptyline Are Substrates and Inhibitors of the AcrB Multidrug Efflux Pump.mBio. 2020 Jun 2;11(3):e00465-20. doi: 10.1128/mBio.00465-20. mBio. 2020. PMID: 32487753 Free PMC article.
-
Tripartite efflux pumps of the RND superfamily: what did we learn from computational studies?Microbiology (Reading). 2023 Mar;169(3):001307. doi: 10.1099/mic.0.001307. Microbiology (Reading). 2023. PMID: 36972322 Free PMC article. Review.
-
Perturbed structural dynamics underlie inhibition and altered efflux of the multidrug resistance pump AcrB.Nat Commun. 2020 Nov 4;11(1):5565. doi: 10.1038/s41467-020-19397-2. Nat Commun. 2020. PMID: 33149158 Free PMC article.
-
Structure, Assembly, and Function of Tripartite Efflux and Type 1 Secretion Systems in Gram-Negative Bacteria.Chem Rev. 2021 May 12;121(9):5479-5596. doi: 10.1021/acs.chemrev.1c00055. Epub 2021 Apr 28. Chem Rev. 2021. PMID: 33909410 Free PMC article. Review.
-
A genetic platform to investigate the functions of bacterial drug efflux pumps.Nat Chem Biol. 2022 Dec;18(12):1399-1409. doi: 10.1038/s41589-022-01119-y. Epub 2022 Sep 5. Nat Chem Biol. 2022. PMID: 36065018
References
-
- World Health Organization. Antimicrobial resistance: global report on surveillance. http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf (January 2016) (2014).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

