Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis
- PMID: 28808260
- PMCID: PMC5556005
- DOI: 10.1038/s41598-017-08603-9
Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis
Abstract
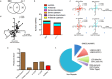
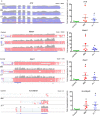
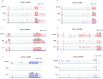
Myositis is characterised by muscle inflammation and weakness. Although generally thought to be driven by a systemic autoimmune response, increasing evidence suggests that intrinsic changes in the muscle might also contribute to the pathogenesis. Long non-coding RNAs (lncRNAs) are a family of novel genes that regulate gene transcription and translation. To determine the potential role of lncRNAs, we employed next generation sequencing to examine the transcriptome in muscle biopsies obtained from two histologically distinct patient populations, inclusion body myositis (IBM) and anti-Jo-1-associated myositis (Jo-1). 1287 mRNAs and 1068 mRNAs were differentially expressed in the muscle from Jo-1 and IBM patients, respectively. Pathway analysis showed the top canonical pathway in both Jo-1 and IBM was oxidative phosphorylation and mitochondrial dysfunction. We identified 731 known and 325 novel lncRNAs in the muscles biopsies. Comparison with controls showed 55 and 46 lncRNAs were differentially expressed in IBM and Jo-1 myositis, respectively. Of these, 16 lncRNAs were differentially expressed in both IBM and Jo-1 myositis and included upregulated H19, lncMyoD and MALAT1. Given that these are known to regulate muscle proliferation and differentiation, we speculate that changes in lncRNAs might contribute to the phenotypic changes in Jo-1 and IBM myositis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Comprehensive transcriptomic analysis shows disturbed calcium homeostasis and deregulation of T lymphocyte apoptosis in inclusion body myositis.J Neurol. 2022 Aug;269(8):4161-4173. doi: 10.1007/s00415-022-11029-7. Epub 2022 Mar 2. J Neurol. 2022. PMID: 35237874 Free PMC article.
-
Muscle Transcriptomics Shows Overexpression of Cadherin 1 in Inclusion Body Myositis.Ann Neurol. 2022 Mar;91(3):317-328. doi: 10.1002/ana.26304. Epub 2022 Feb 11. Ann Neurol. 2022. PMID: 35064929 Free PMC article.
-
Calcium dysregulation, functional calpainopathy, and endoplasmic reticulum stress in sporadic inclusion body myositis.Acta Neuropathol Commun. 2017 Mar 22;5(1):24. doi: 10.1186/s40478-017-0427-7. Acta Neuropathol Commun. 2017. PMID: 28330496 Free PMC article.
-
Long non-coding RNAs: a new frontier in the study of human diseases.Cancer Lett. 2013 Oct 10;339(2):159-66. doi: 10.1016/j.canlet.2013.06.013. Epub 2013 Jun 18. Cancer Lett. 2013. PMID: 23791884 Review.
-
Inclusion body myositis: genetic factors, aberrant protein expression, and autoimmunity.Curr Opin Rheumatol. 2001 Nov;13(6):469-75. doi: 10.1097/00002281-200111000-00003. Curr Opin Rheumatol. 2001. PMID: 11698722 Review.
Cited by
-
Comprehensive transcriptomic analysis shows disturbed calcium homeostasis and deregulation of T lymphocyte apoptosis in inclusion body myositis.J Neurol. 2022 Aug;269(8):4161-4173. doi: 10.1007/s00415-022-11029-7. Epub 2022 Mar 2. J Neurol. 2022. PMID: 35237874 Free PMC article.
-
Inclusion body myositis, viral infections, and TDP-43: a narrative review.Clin Exp Med. 2024 May 2;24(1):91. doi: 10.1007/s10238-024-01353-9. Clin Exp Med. 2024. PMID: 38693436 Free PMC article. Review.
-
Involvement of long noncoding RNAs in the pathogenesis of autoimmune diseases.J Transl Autoimmun. 2020 Mar 17;3:100044. doi: 10.1016/j.jtauto.2020.100044. eCollection 2020. J Transl Autoimmun. 2020. PMID: 32743525 Free PMC article. Review.
-
Long non-coding RNA H19, a novel therapeutic target for pancreatic cancer.Mol Med. 2020 Apr 9;26(1):30. doi: 10.1186/s10020-020-00156-4. Mol Med. 2020. PMID: 32272875 Free PMC article. Review.
-
Sporadic Inclusion Body Myositis: An Acquired Mitochondrial Disease with Extras.Biomolecules. 2019 Jan 7;9(1):15. doi: 10.3390/biom9010015. Biomolecules. 2019. PMID: 30621041 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

