Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression
- PMID: 28806168
- PMCID: PMC5555720
- DOI: 10.7554/eLife.25125
Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression
Abstract
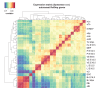
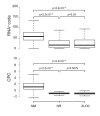
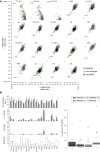
To determine the dynamics of allelic-specific expression during mouse development, we analyzed RNA-seq data from 23 F1 tissues from different developmental stages, including 19 female tissues allowing X chromosome inactivation (XCI) escapers to also be detected. We demonstrate that allelic expression arising from genetic or epigenetic differences is highly tissue-specific. We find that tissue-specific strain-biased gene expression may be regulated by tissue-specific enhancers or by post-transcriptional differences in stability between the alleles. We also find that escape from X-inactivation is tissue-specific, with leg muscle showing an unexpectedly high rate of XCI escapers. By surveying a range of tissues during development, and performing extensive validation, we are able to provide a high confidence list of mouse imprinted genes including 18 novel genes. This shows that cluster size varies dynamically during development and can be substantially larger than previously thought, with the Igf2r cluster extending over 10 Mb in placenta.
Keywords: H3K27ac; XCI escapers; allelic expression; developmental biology; enhancer; evolutionary biology; genomic imprinting; genomics; mouse; stem cells; strain-biased expression.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures








Similar articles
-
X-Chromosome Inactivation and Escape from X Inactivation in Mouse.Methods Mol Biol. 2018;1861:205-219. doi: 10.1007/978-1-4939-8766-5_15. Methods Mol Biol. 2018. PMID: 30218369 Free PMC article.
-
Paternally biased X inactivation in mouse neonatal brain.Genome Biol. 2010;11(7):R79. doi: 10.1186/gb-2010-11-7-r79. Epub 2010 Jul 27. Genome Biol. 2010. PMID: 20663224 Free PMC article.
-
Allelome.PRO, a pipeline to define allele-specific genomic features from high-throughput sequencing data.Nucleic Acids Res. 2015 Dec 2;43(21):e146. doi: 10.1093/nar/gkv727. Epub 2015 Jul 21. Nucleic Acids Res. 2015. PMID: 26202974 Free PMC article.
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
X-tra! X-tra! News from the mouse X chromosome.Dev Biol. 2006 Oct 15;298(2):344-53. doi: 10.1016/j.ydbio.2006.07.011. Epub 2006 Jul 15. Dev Biol. 2006. PMID: 16916508 Review.
Cited by
-
Simultaneous brain cell type and lineage determined by scRNA-seq reveals stereotyped cortical development.Cell Syst. 2022 Jun 15;13(6):438-453.e5. doi: 10.1016/j.cels.2022.03.006. Epub 2022 Apr 21. Cell Syst. 2022. PMID: 35452605 Free PMC article.
-
Systematic Analysis of Monoallelic Gene Expression and Chromatin Accessibility Across Multiple Tissues in Hybrid Mice.Front Cell Dev Biol. 2021 Sep 23;9:717555. doi: 10.3389/fcell.2021.717555. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34631706 Free PMC article.
-
Allele-specific RNA-seq expression profiling of imprinted genes in mouse isogenic pluripotent states.Epigenetics Chromatin. 2019 Feb 15;12(1):14. doi: 10.1186/s13072-019-0259-8. Epigenetics Chromatin. 2019. PMID: 30767785 Free PMC article.
-
Imprinted Grb10, encoding growth factor receptor bound protein 10, regulates fetal growth independently of the insulin-like growth factor type 1 receptor (Igf1r) and insulin receptor (Insr) genes.BMC Biol. 2024 May 30;22(1):127. doi: 10.1186/s12915-024-01926-w. BMC Biol. 2024. PMID: 38816743 Free PMC article.
-
Trans- and cis-acting effects of Firre on epigenetic features of the inactive X chromosome.Nat Commun. 2020 Nov 27;11(1):6053. doi: 10.1038/s41467-020-19879-3. Nat Commun. 2020. PMID: 33247132 Free PMC article.
References
-
- Andergassen D, Dotter CP, Kulinski TM, Guenzl PM, Bammer PC, Barlow DP, Pauler FM, Hudson QJ. Allelome.pro, a pipeline to define allele-specific genomic features from high-throughput sequencing data. Nucleic Acids Research. 2015;43:e146. doi: 10.1093/nar/gkv727. - DOI - PMC - PubMed
-
- Baran Y, Subramaniam M, Biton A, Tukiainen T, Tsang EK, Rivas MA, Pirinen M, Gutierrez-Arcelus M, Smith KS, Kukurba KR, Zhang R, Eng C, Torgerson DG, Urbanek C, Li JB, Rodriguez-Santana JR, Burchard EG, Seibold MA, MacArthur DG, Montgomery SB, Zaitlen NA, Lappalainen T, GTEx Consortium The landscape of genomic imprinting across diverse adult human tissues. Genome Research. 2015;25:927–936. doi: 10.1101/gr.192278.115. - DOI - PMC - PubMed
-
- Behringer RR, Gertsenstein M, Nagy K, V, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2014.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

