Rab44, a novel large Rab GTPase, negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation
- PMID: 28791425
- PMCID: PMC11105776
- DOI: 10.1007/s00018-017-2607-9
Rab44, a novel large Rab GTPase, negatively regulates osteoclast differentiation by modulating intracellular calcium levels followed by NFATc1 activation
Abstract
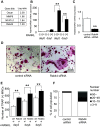
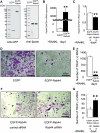
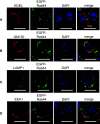
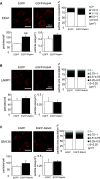
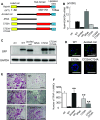
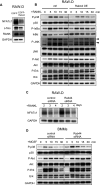
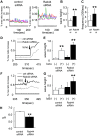
Rab44 is an atypical Rab GTPase that contains some additional domains such as the EF-hand and coiled-coil domains as well as Rab-GTPase domain. Although Rab44 genes have been found in mammalian genomes, no studies concerning Rab44 have been reported yet. Here, we identified Rab44 as an upregulated protein during osteoclast differentiation. Knockdown of Rab44 by small interfering RNA promotes RANKL-induced osteoclast differentiation of the murine monocytic cell line, RAW-D or of bone marrow-derived macrophages (BMMs). In contrast, overexpression of Rab44 prevents osteoclast differentiation. Rab44 was localized in the Golgi complex and lysosomes, and Rab44 overexpression caused an enlargement of early endosomes. A series of deletion mutant studies of Rab44 showed that the coiled-coil domain and lipidation sites of Rab44 is important for regulation of osteoclast differentiation. Mechanistically, Rab44 affects nuclear factor of activated T-cells c1 (NFATc1) signaling in RANKL-stimulated macrophages. Moreover, Rab44 depletion caused an elevation in intracellular Ca2+ transients upon RANKL stimulation, and particularly regulated lysosomal Ca2+ influx. Taken together, these results suggest that Rab44 negatively regulates osteoclast differentiation by modulating intracellular Ca2+ levels followed by NFATc1 activation.
Keywords: Intracellular Ca2+ levels; NFATc1; Osteoclast; Rab GTPase; Rab44.
Figures
Similar articles
-
Large Rab GTPases: Novel Membrane Trafficking Regulators with a Calcium Sensor and Functional Domains.Int J Mol Sci. 2021 Jul 19;22(14):7691. doi: 10.3390/ijms22147691. Int J Mol Sci. 2021. PMID: 34299309 Free PMC article. Review.
-
Rab11A Functions as a Negative Regulator of Osteoclastogenesis through Dictating Lysosome-Induced Proteolysis of c-fms and RANK Surface Receptors.Cells. 2020 Oct 31;9(11):2384. doi: 10.3390/cells9112384. Cells. 2020. PMID: 33142674 Free PMC article.
-
Expression and localisation of Rab44 in immune-related cells change during cell differentiation and stimulation.Sci Rep. 2020 Jul 1;10(1):10728. doi: 10.1038/s41598-020-67638-7. Sci Rep. 2020. PMID: 32612275 Free PMC article.
-
Coronin1C Is a GDP-Specific Rab44 Effector That Controls Osteoclast Formation by Regulating Cell Motility in Macrophages.Int J Mol Sci. 2022 Jun 14;23(12):6619. doi: 10.3390/ijms23126619. Int J Mol Sci. 2022. PMID: 35743062 Free PMC article.
-
Homer2 and Homer3 modulate RANKL-induced NFATc1 signaling in osteoclastogenesis and bone metabolism.J Endocrinol. 2019 Sep;242(3):241-249. doi: 10.1530/JOE-19-0123. J Endocrinol. 2019. PMID: 31319381 Free PMC article.
Cited by
-
Rab44 isoforms similarly promote lysosomal exocytosis, but exhibit differential localization in mast cells.FEBS Open Bio. 2021 Apr;11(4):1165-1185. doi: 10.1002/2211-5463.13133. Epub 2021 Mar 19. FEBS Open Bio. 2021. PMID: 33641252 Free PMC article.
-
Large Rab GTPases: Novel Membrane Trafficking Regulators with a Calcium Sensor and Functional Domains.Int J Mol Sci. 2021 Jul 19;22(14):7691. doi: 10.3390/ijms22147691. Int J Mol Sci. 2021. PMID: 34299309 Free PMC article. Review.
-
Rab11A Functions as a Negative Regulator of Osteoclastogenesis through Dictating Lysosome-Induced Proteolysis of c-fms and RANK Surface Receptors.Cells. 2020 Oct 31;9(11):2384. doi: 10.3390/cells9112384. Cells. 2020. PMID: 33142674 Free PMC article.
-
The Role of Rab GTPases in the development of genetic and malignant diseases.Mol Cell Biochem. 2024 Feb;479(2):255-281. doi: 10.1007/s11010-023-04727-x. Epub 2023 Apr 15. Mol Cell Biochem. 2024. PMID: 37060515 Review.
-
Dennd2c Negatively Controls Multinucleation and Differentiation in Osteoclasts by Regulating Actin Polymerization and Protrusion Formation.Int J Mol Sci. 2024 Oct 25;25(21):11479. doi: 10.3390/ijms252111479. Int J Mol Sci. 2024. PMID: 39519032 Free PMC article.
References
-
- Surkont J, Diekmann Y, Pereira-Leal JB. Rabifier2: an improved bioinformatic classifier of Rab GTPases. Bioinformatics (Oxford, England) 2016 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

