Massively parallel unsupervised single-particle cryo-EM data clustering via statistical manifold learning
- PMID: 28786986
- PMCID: PMC5546606
- DOI: 10.1371/journal.pone.0182130
Massively parallel unsupervised single-particle cryo-EM data clustering via statistical manifold learning
Abstract
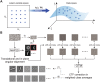
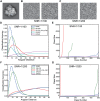
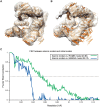
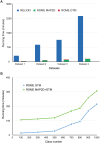
Structural heterogeneity in single-particle cryo-electron microscopy (cryo-EM) data represents a major challenge for high-resolution structure determination. Unsupervised classification may serve as the first step in the assessment of structural heterogeneity. However, traditional algorithms for unsupervised classification, such as K-means clustering and maximum likelihood optimization, may classify images into wrong classes with decreasing signal-to-noise-ratio (SNR) in the image data, yet demand increased computational costs. Overcoming these limitations requires further development of clustering algorithms for high-performance cryo-EM data processing. Here we introduce an unsupervised single-particle clustering algorithm derived from a statistical manifold learning framework called generative topographic mapping (GTM). We show that unsupervised GTM clustering improves classification accuracy by about 40% in the absence of input references for data with lower SNRs. Applications to several experimental datasets suggest that our algorithm can detect subtle structural differences among classes via a hierarchical clustering strategy. After code optimization over a high-performance computing (HPC) environment, our software implementation was able to generate thousands of reference-free class averages within hours in a massively parallel fashion, which allows a significant improvement on ab initio 3D reconstruction and assists in the computational purification of homogeneous datasets for high-resolution visualization.
Conflict of interest statement
Figures



Similar articles
-
An Unsupervised Classification Algorithm for Heterogeneous Cryo-EM Projection Images Based on Autoencoders.Int J Mol Sci. 2023 May 6;24(9):8380. doi: 10.3390/ijms24098380. Int J Mol Sci. 2023. PMID: 37176089 Free PMC article.
-
Unsupervised Cryo-EM Data Clustering through Adaptively Constrained K-Means Algorithm.PLoS One. 2016 Dec 13;11(12):e0167765. doi: 10.1371/journal.pone.0167765. eCollection 2016. PLoS One. 2016. PMID: 27959895 Free PMC article.
-
AutoCryoPicker: an unsupervised learning approach for fully automated single particle picking in Cryo-EM images.BMC Bioinformatics. 2019 Jun 13;20(1):326. doi: 10.1186/s12859-019-2926-y. BMC Bioinformatics. 2019. PMID: 31195977 Free PMC article.
-
Processing of Structurally Heterogeneous Cryo-EM Data in RELION.Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review.
-
Cryo-Electron Tomography and Subtomogram Averaging.Methods Enzymol. 2016;579:329-67. doi: 10.1016/bs.mie.2016.04.014. Epub 2016 Jun 22. Methods Enzymol. 2016. PMID: 27572733 Review.
Cited by
-
New software tools in EMAN2 inspired by EMDatabank map challenge.J Struct Biol. 2018 Nov;204(2):283-290. doi: 10.1016/j.jsb.2018.09.002. Epub 2018 Sep 4. J Struct Biol. 2018. PMID: 30189321 Free PMC article.
-
Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.Nat Commun. 2018 Apr 10;9(1):1360. doi: 10.1038/s41467-018-03785-w. Nat Commun. 2018. PMID: 29636472 Free PMC article.
-
CryoEM structure of Drosophila flight muscle thick filaments at 7 Å resolution.Life Sci Alliance. 2020 Jul 27;3(8):e202000823. doi: 10.26508/lsa.202000823. Print 2020 Aug. Life Sci Alliance. 2020. PMID: 32718994 Free PMC article.
-
An Unsupervised Classification Algorithm for Heterogeneous Cryo-EM Projection Images Based on Autoencoders.Int J Mol Sci. 2023 May 6;24(9):8380. doi: 10.3390/ijms24098380. Int J Mol Sci. 2023. PMID: 37176089 Free PMC article.
-
USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.Nature. 2022 May;605(7910):567-574. doi: 10.1038/s41586-022-04671-8. Epub 2022 Apr 27. Nature. 2022. PMID: 35477760 Free PMC article.
References
-
- Frank J. Three-dimensional electron microscopy of macromolecular assemblies: visualization of biological molecules in their native state: Oxford University Press; 2006.
-
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature methods. 2007;4(1):27–9. doi: 10.1038/nmeth992 - DOI - PubMed
-
- Katsevich E, Katsevich A, Singer A. Covariance matrix estimation for the cryo-EM heterogeneity problem. SIAM journal on imaging sciences. 2015;8(1):126–85. doi: 10.1137/130935434 - DOI - PMC - PubMed
-
- Schwander P, Fung R, Ourmazd A. Conformations of macromolecules and their complexes from heterogeneous datasets. Phil Trans R Soc B. 2014;369(1647):20130567 doi: 10.1098/rstb.2013.0567 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

