HIV virions sense plasma membrane heterogeneity for cell entry
- PMID: 28782011
- PMCID: PMC5489272
- DOI: 10.1126/sciadv.1700338
HIV virions sense plasma membrane heterogeneity for cell entry
Abstract
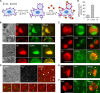
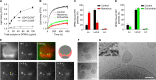
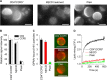
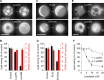
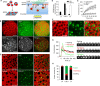
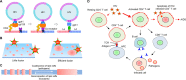
It has been proposed that cholesterol in host cell membranes plays a pivotal role for cell entry of HIV. However, it remains largely unknown why virions prefer cholesterol-rich heterogeneous membranes to uniformly fluid membranes for membrane fusion. Using giant plasma membrane vesicles containing cholesterol-rich ordered and cholesterol-poor fluid lipid domains, we demonstrate that the HIV receptor CD4 is substantially sequestered into ordered domains, whereas the co-receptor CCR5 localizes preferentially at ordered/disordered domain boundaries. We also show that HIV does not fuse from within ordered regions of the plasma membrane but rather at their boundaries. Ordered/disordered lipid domain coexistence is not required for HIV attachment but is a prerequisite for successful fusion. We propose that HIV virions sense and exploit membrane discontinuities to gain entry into cells. This study provides surprising answers to the long-standing question about the roles of cholesterol and ordered lipid domains in cell entry of HIV and perhaps other enveloped viruses.
Keywords: HIV; cell entry; cholesterol; membrane domain; membrane fusion.
Figures
Similar articles
-
HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains.Nat Chem Biol. 2015 Jun;11(6):424-31. doi: 10.1038/nchembio.1800. Epub 2015 Apr 27. Nat Chem Biol. 2015. PMID: 25915200 Free PMC article.
-
Membrane organization of virus and target cell plays a role in HIV entry.Biochimie. 2014 Dec;107 Pt A:22-7. doi: 10.1016/j.biochi.2014.08.015. Epub 2014 Sep 1. Biochimie. 2014. PMID: 25193376 Free PMC article. Review.
-
Quantifying the Sensitivity of HIV-1 Viral Entry to Receptor and Coreceptor Expression.J Phys Chem B. 2016 Jul 7;120(26):6189-99. doi: 10.1021/acs.jpcb.6b02102. Epub 2016 May 20. J Phys Chem B. 2016. PMID: 27137677
-
Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells.J Virol. 2002 May;76(10):4709-22. doi: 10.1128/jvi.76.10.4709-4722.2002. J Virol. 2002. PMID: 11967288 Free PMC article.
-
The HIV-1 Entry Process: A Stoichiometric View.Trends Microbiol. 2015 Dec;23(12):763-774. doi: 10.1016/j.tim.2015.09.003. Epub 2015 Nov 2. Trends Microbiol. 2015. PMID: 26541228 Review.
Cited by
-
Fluorescence Microscopy of the HIV-1 Envelope.Viruses. 2020 Mar 21;12(3):348. doi: 10.3390/v12030348. Viruses. 2020. PMID: 32245254 Free PMC article. Review.
-
A novel in vitro system of supported planar endosomal membranes (SPEMs) reveals an enhancing role for cathepsin B in the final stage of Ebola virus fusion and entry.Microbiol Spectr. 2023 Sep 20;11(5):e0190823. doi: 10.1128/spectrum.01908-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37728342 Free PMC article.
-
Imaging Viral Infection by Fluorescence Microscopy: Focus on HIV-1 Early Stage.Viruses. 2021 Jan 30;13(2):213. doi: 10.3390/v13020213. Viruses. 2021. PMID: 33573241 Free PMC article. Review.
-
Barcoding Biological Reactions with DNA-Functionalized Vesicles.Angew Chem Int Ed Engl. 2019 Dec 16;58(51):18683-18690. doi: 10.1002/anie.201911544. Epub 2019 Oct 31. Angew Chem Int Ed Engl. 2019. PMID: 31596992 Free PMC article. Review.
-
Lipid Rafts: Controversies Resolved, Mysteries Remain.Trends Cell Biol. 2020 May;30(5):341-353. doi: 10.1016/j.tcb.2020.01.009. Epub 2020 Feb 20. Trends Cell Biol. 2020. PMID: 32302547 Free PMC article. Review.
References
-
- Simons K., Ikonen E., Functional rafts in cell membranes. Nature 387, 569–572 (1997). - PubMed
-
- Lingwood D., Kaiser H.-J., Levental I., Simons K., Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 37, 955–960 (2009). - PubMed
-
- Brown D. A., London E., Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14, 111–136 (1998). - PubMed
-
- Anderson R. G. W., Jacobson K., A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296, 1821–1825 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

