Solution NMR structure of the TRIM21 B-box2 and identification of residues involved in its interaction with the RING domain
- PMID: 28753623
- PMCID: PMC5533445
- DOI: 10.1371/journal.pone.0181551
Solution NMR structure of the TRIM21 B-box2 and identification of residues involved in its interaction with the RING domain
Abstract
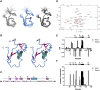
Tripartite motif-containing (TRIM) proteins are defined by the sequential arrangement of RING, B-box and coiled-coil domains (RBCC), where the B-box domain is a unique feature of the TRIM protein family. TRIM21 is an E3 ubiquitin-protein ligase implicated in innate immune signaling by acting as an autoantigen and by modifying interferon regulatory factors. Here we report the three-dimensional solution structure of the TRIM21 B-box2 domain by nuclear magnetic resonance (NMR) spectroscopy. The structure of the B-box2 domain, comprising TRIM21 residues 86-130, consists of a short α-helical segment with an N-terminal short β-strand and two anti-parallel β-strands jointly found the core, and adopts a RING-like fold. This ββαβ core largely defines the overall fold of the TRIM21 B-box2 and the coordination of one Zn2+ ion stabilizes the tertiary structure of the protein. Using NMR titration experiments, we have identified an exposed interaction surface, a novel interaction patch where the B-box2 is likely to bind the N-terminal RING domain. Our structure together with comparisons with other TRIM B-box domains jointly reveal how its different surfaces are employed for various modular interactions, and provides extended understanding of how this domain relates to flanking domains in TRIM proteins.
Conflict of interest statement
Figures
Similar articles
-
Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold.J Mol Biol. 2007 May 25;369(1):1-10. doi: 10.1016/j.jmb.2007.03.017. Epub 2007 Mar 15. J Mol Biol. 2007. PMID: 17428496
-
Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING.J Mol Biol. 2006 Apr 28;358(2):532-45. doi: 10.1016/j.jmb.2006.02.009. Epub 2006 Feb 20. J Mol Biol. 2006. PMID: 16529770
-
Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers.Biochemistry. 2008 Feb 26;47(8):2450-7. doi: 10.1021/bi7018496. Epub 2008 Jan 26. Biochemistry. 2008. PMID: 18220417
-
Multiple Roles of TRIM21 in Virus Infection.Int J Mol Sci. 2023 Jan 14;24(2):1683. doi: 10.3390/ijms24021683. Int J Mol Sci. 2023. PMID: 36675197 Free PMC article. Review.
-
Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases.Biochem Soc Trans. 2020 Dec 18;48(6):2615-2624. doi: 10.1042/BST20200383. Biochem Soc Trans. 2020. PMID: 33170204 Free PMC article. Review.
Cited by
-
Tripartite Motif-Containing Protein 65 (TRIM65) Inhibits Hepatitis B Virus Transcription.Viruses. 2024 May 31;16(6):890. doi: 10.3390/v16060890. Viruses. 2024. PMID: 38932182 Free PMC article.
-
Roles of TRIM21/Ro52 in connective tissue disease-associated interstitial lung diseases.Front Immunol. 2024 Aug 6;15:1435525. doi: 10.3389/fimmu.2024.1435525. eCollection 2024. Front Immunol. 2024. PMID: 39165359 Free PMC article. Review.
-
TRIM21/Ro52 - Roles in Innate Immunity and Autoimmune Disease.Front Immunol. 2021 Sep 6;12:738473. doi: 10.3389/fimmu.2021.738473. eCollection 2021. Front Immunol. 2021. PMID: 34552597 Free PMC article. Review.
-
A Comparison of Bonded and Nonbonded Zinc(II) Force Fields with NMR Data.Int J Mol Sci. 2023 Mar 13;24(6):5440. doi: 10.3390/ijms24065440. Int J Mol Sci. 2023. PMID: 36982515 Free PMC article.
-
To TRIM or not to TRIM: the balance of host-virus interactions mediated by the ubiquitin system.J Gen Virol. 2019 Dec;100(12):1641-1662. doi: 10.1099/jgv.0.001341. J Gen Virol. 2019. PMID: 31661051 Free PMC article.
References
-
- Hatakeyama S. TRIM proteins and cancer. Nature Publishing Group. Nature Publishing Group; 2011;11: 792–804. doi: 10.1038/nrc3139 - DOI - PubMed
-
- Rajsbaum R, García-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426: 1265–1284. doi: 10.1016/j.jmb.2013.12.005 - DOI - PMC - PubMed
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67: 425–479. doi: 10.1146/annurev.biochem.67.1.425 - DOI - PubMed
-
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3: e1487 doi: 10.1371/journal.pone.0001487 - DOI - PMC - PubMed
-
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21: 301–307. doi: 10.1038/nsmb.2780 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

