Complex chromosomal neighborhood effects determine the adaptive potential of a gene under selection
- PMID: 28738969
- PMCID: PMC5526668
- DOI: 10.7554/eLife.25100
Complex chromosomal neighborhood effects determine the adaptive potential of a gene under selection
Abstract
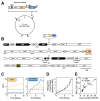
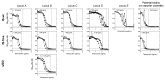
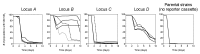
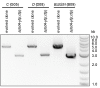
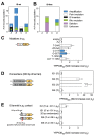
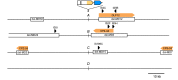
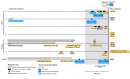
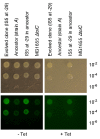
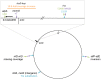
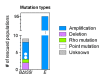
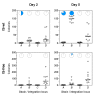
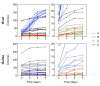
How the organization of genes on a chromosome shapes adaptation is essential for understanding evolutionary paths. Here, we investigate how adaptation to rapidly increasing levels of antibiotic depends on the chromosomal neighborhood of a drug-resistance gene inserted at different positions of the Escherichia coli chromosome. Using a dual-fluorescence reporter that allows us to distinguish gene amplifications from other up-mutations, we track in real-time adaptive changes in expression of the drug-resistance gene. We find that the relative contribution of several mutation types differs systematically between loci due to properties of neighboring genes: essentiality, expression, orientation, termination, and presence of duplicates. These properties determine rate and fitness effects of gene amplification, deletions, and mutations compromising transcriptional termination. Thus, the adaptive potential of a gene under selection is a system-property with a complex genetic basis that is specific for each chromosomal locus, and it can be inferred from detailed functional and genomic data.
Keywords: E. coli; adaptation; bacterial genetics; chromosomal architecture; computational biology; evolutionary biology; experimental evolution; gene expression; genomics; regulatory evolution; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





Similar articles
-
Network models of TEM β-lactamase mutations coevolving under antibiotic selection show modular structure and anticipate evolutionary trajectories.PLoS Comput Biol. 2011 Sep;7(9):e1002184. doi: 10.1371/journal.pcbi.1002184. Epub 2011 Sep 22. PLoS Comput Biol. 2011. PMID: 21966264 Free PMC article.
-
Quantifying the Determinants of Evolutionary Dynamics Leading to Drug Resistance.PLoS Biol. 2015 Nov 18;13(11):e1002299. doi: 10.1371/journal.pbio.1002299. eCollection 2015. PLoS Biol. 2015. PMID: 26581035 Free PMC article.
-
Genomics of rapid adaptation to antibiotics: convergent evolution and scalable sequence amplification.Genome Biol Evol. 2014 May 20;6(6):1287-301. doi: 10.1093/gbe/evu106. Genome Biol Evol. 2014. PMID: 24850796 Free PMC article.
-
Escherichia coli evolution during stationary phase.Res Microbiol. 2004 Jun;155(5):328-36. doi: 10.1016/j.resmic.2004.01.014. Res Microbiol. 2004. PMID: 15207864 Review.
-
[Analysis of Drug Resistance Using Experimental Evolution].Yakugaku Zasshi. 2017;137(4):373-376. doi: 10.1248/yakushi.16-00235-1. Yakugaku Zasshi. 2017. PMID: 28381708 Review. Japanese.
Cited by
-
Leaky resistance and the conditions for the existence of lytic bacteriophage.PLoS Biol. 2018 Aug 16;16(8):e2005971. doi: 10.1371/journal.pbio.2005971. eCollection 2018 Aug. PLoS Biol. 2018. PMID: 30114198 Free PMC article.
-
Single-cell copy number variant detection reveals the dynamics and diversity of adaptation.PLoS Biol. 2018 Dec 18;16(12):e3000069. doi: 10.1371/journal.pbio.3000069. eCollection 2018 Dec. PLoS Biol. 2018. PMID: 30562346 Free PMC article.
-
Living Organisms Author Their Read-Write Genomes in Evolution.Biology (Basel). 2017 Dec 6;6(4):42. doi: 10.3390/biology6040042. Biology (Basel). 2017. PMID: 29211049 Free PMC article. Review.
-
Local genetic context shapes the function of a gene regulatory network.Elife. 2021 Mar 8;10:e65993. doi: 10.7554/eLife.65993. Elife. 2021. PMID: 33683203 Free PMC article.
-
Gene amplification mutations originate prior to selective stress in Acinetobacter baylyi.G3 (Bethesda). 2023 Mar 9;13(3):jkac327. doi: 10.1093/g3journal/jkac327. G3 (Bethesda). 2023. PMID: 36504387 Free PMC article.
References
-
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LF, van Lohuizen M, van Steensel B, Jong Jde, Ridder Jde, Lohuizen Mvan, Steensel Bvan. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

