Functionality of tRNAs encoded in a mobile genetic element from an acidophilic bacterium
- PMID: 28708455
- PMCID: PMC6103686
- DOI: 10.1080/15476286.2017.1349049
Functionality of tRNAs encoded in a mobile genetic element from an acidophilic bacterium
Abstract
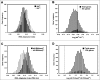
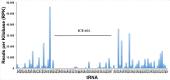
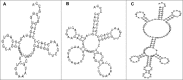
The genome of the acidophilic, bioleaching bacterium Acidithiobacillus ferrooxidans, strain ATCC 23270, contains 95 predicted tRNA genes. Thirty-six of these genes (all 20 species) are clustered within an actively excising integrative-conjugative element (ICEAfe1). We speculated that these tRNA genes might have a role in adapting the bacterial tRNA pool to the codon usage of ICEAfe1 genes. To answer this question, we performed theoretical calculations of the global tRNA adaptation index to the entire A. ferrooxidans genome with and without the ICEAfe1 encoded tRNA genes. Based on these calculations, we observed that tRNAs encoded in ICEAfe1 negatively contribute to adapt the tRNA pool to the codon use in A. ferrooxidans. Although some of the tRNAs encoded in ICEAfe1 are functional in aminoacylation or protein synthesis, we found that they are expressed at low levels. These findings, along with the identification of a tRNA-like RNA encoded in the same cluster, led us to speculate that tRNA genes encoded in the mobile genetic element ICEAfe1 might have acquired mutations that would result in either inactivation or the acquisition of new functions.
Keywords: Transfer RNA; acidophilic bacterium; horizontal gene transfer; mobile genetic elements; phylogeny; translation.
Figures


Similar articles
-
ICE Afe 1, an actively excising genetic element from the biomining bacterium Acidithiobacillus ferrooxidans.J Mol Microbiol Biotechnol. 2012;22(6):399-407. doi: 10.1159/000346669. Epub 2013 Mar 7. J Mol Microbiol Biotechnol. 2012. PMID: 23486178
-
A DNA segment encoding the anticodon stem/loop of tRNA determines the specific recombination of integrative-conjugative elements in Acidithiobacillus species.RNA Biol. 2018;15(4-5):492-499. doi: 10.1080/15476286.2017.1408765. Epub 2017 Dec 20. RNA Biol. 2018. PMID: 29168417 Free PMC article.
-
Toxin-antitoxin systems in the mobile genome of Acidithiobacillus ferrooxidans.PLoS One. 2014 Nov 10;9(11):e112226. doi: 10.1371/journal.pone.0112226. eCollection 2014. PLoS One. 2014. PMID: 25384039 Free PMC article.
-
Genomic insights into the iron uptake mechanisms of the biomining microorganism Acidithiobacillus ferrooxidans.J Ind Microbiol Biotechnol. 2005 Dec;32(11-12):606-14. doi: 10.1007/s10295-005-0233-2. Epub 2005 May 14. J Ind Microbiol Biotechnol. 2005. PMID: 15895264 Review.
-
tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations.Biochimie. 2005 Sep-Oct;87(9-10):835-45. doi: 10.1016/j.biochi.2005.02.014. Biochimie. 2005. PMID: 15925436 Review.
Cited by
-
Beyond the Limits: tRNA Array Units in Mycobacterium Genomes.Front Microbiol. 2018 May 17;9:1042. doi: 10.3389/fmicb.2018.01042. eCollection 2018. Front Microbiol. 2018. PMID: 29867913 Free PMC article.
-
The Mobilome-Enriched Genome of the Competence-Deficient Streptococcus pneumoniae BM6001, the Original Host of Integrative Conjugative Element Tn5253, Is Phylogenetically Distinct from Historical Pneumococcal Genomes.Microorganisms. 2023 Jun 23;11(7):1646. doi: 10.3390/microorganisms11071646. Microorganisms. 2023. PMID: 37512819 Free PMC article.
-
Mycobacterium genus and tRNA arrays.Mem Inst Oswaldo Cruz. 2019;114:e180443. doi: 10.1590/0074-02760180443. Epub 2019 May 13. Mem Inst Oswaldo Cruz. 2019. PMID: 31090860 Free PMC article.
-
Role of a cryptic tRNA gene operon in survival under translational stress.Nucleic Acids Res. 2021 Sep 7;49(15):8757-8776. doi: 10.1093/nar/gkab661. Nucleic Acids Res. 2021. PMID: 34379789 Free PMC article.
-
Comparative Genomics Suggests Mechanisms of Genetic Adaptation toward the Catabolism of the Phenylurea Herbicide Linuron in Variovorax.Genome Biol Evol. 2020 Jun 1;12(6):827-841. doi: 10.1093/gbe/evaa085. Genome Biol Evol. 2020. PMID: 32359160 Free PMC article.
References
-
- Giegé R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol 2008; 15:1007-14; PMID:18836497; https://doi.org/10.1038/nsmb.1498 - DOI - PubMed
-
- Ibba M, Söll D. Aminoacyl-tRNA Synthesis. Annu Rev Biochem 2000; 69:617-50; PMID:10966471; https://doi.org/10.1146/annurev.biochem.69.1.617 - DOI - PubMed
-
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 2009; 37:D93-7; PMID:18984615; https://doi.org/10.1093/nar/gkn787 - DOI - PMC - PubMed
-
- Bermudez-Santana C, Attolini C, Kirsten T, Engelhardt J, Prohaska SJ, Steigele S, Stadler PF. Genomic organization of eukaryotic tRNAs. BMC Genomics 2010; 11:270; PMID:20426822; https://doi.org/10.1186/1471-2164-11-270 - DOI - PMC - PubMed
-
- Gingold H, Dahan O, Pilpel Y. Dynamic changes in translational efficiency are deduced from codon usage of the transcriptome. Nucleic Acids Res 2012; 40:10053-63; PMID:22941644; https://doi.org/10.1093/nar/gks772 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
