Precarious maintenance of simple DNA repeats in eukaryotes
- PMID: 28703879
- PMCID: PMC5577815
- DOI: 10.1002/bies.201700077
Precarious maintenance of simple DNA repeats in eukaryotes
Abstract
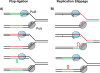
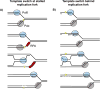
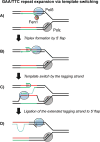
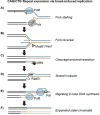
In this review, we discuss how two evolutionarily conserved pathways at the interface of DNA replication and repair, template switching and break-induced replication, lead to the deleterious large-scale expansion of trinucleotide DNA repeats that cause numerous hereditary diseases. We highlight that these pathways, which originated in prokaryotes, may be subsequently hijacked to maintain long DNA microsatellites in eukaryotes. We suggest that the negative mutagenic outcomes of these pathways, exemplified by repeat expansion diseases, are likely outweighed by their positive role in maintaining functional repetitive regions of the genome such as telomeres and centromeres.
Keywords: DNA microsatellites; break-induced replication; centromeres; replication fork stalling; telomeres; template switching; trinucleotide repeats.
© 2017 WILEY Periodicals, Inc.
Figures

Similar articles
-
Restarted replication forks are error-prone and cause CAG repeat expansions and contractions.PLoS Genet. 2021 Oct 21;17(10):e1009863. doi: 10.1371/journal.pgen.1009863. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34673780 Free PMC article.
-
Metabolism of DNA secondary structures at the eukaryotic replication fork.DNA Repair (Amst). 2014 Jul;19:152-62. doi: 10.1016/j.dnarep.2014.03.016. Epub 2014 May 9. DNA Repair (Amst). 2014. PMID: 24815912
-
Replication stalling and heteroduplex formation within CAG/CTG trinucleotide repeats by mismatch repair.DNA Repair (Amst). 2016 Jun;42:94-106. doi: 10.1016/j.dnarep.2016.03.002. Epub 2016 Mar 16. DNA Repair (Amst). 2016. PMID: 27045900
-
Break-induced replication links microsatellite expansion to complex genome rearrangements.Bioessays. 2017 Aug;39(8):10.1002/bies.201700025. doi: 10.1002/bies.201700025. Epub 2017 Jun 16. Bioessays. 2017. PMID: 28621832 Free PMC article. Review.
-
Microsatellite and trinucleotide-repeat evolution: evidence for mutational bias and different rates of evolution in different lineages.Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1095-9. doi: 10.1098/rstb.1999.0465. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434312 Free PMC article. Review.
Cited by
-
Mutation and selection processes regulating short tandem repeats give rise to genetic and phenotypic diversity across species.J Evol Biol. 2023 Feb;36(2):321-336. doi: 10.1111/jeb.14106. Epub 2022 Oct 26. J Evol Biol. 2023. PMID: 36289560 Free PMC article. Review.
-
Restarted replication forks are error-prone and cause CAG repeat expansions and contractions.PLoS Genet. 2021 Oct 21;17(10):e1009863. doi: 10.1371/journal.pgen.1009863. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34673780 Free PMC article.
-
p53 Binds Preferentially to Non-B DNA Structures Formed by the Pyrimidine-Rich Strands of GAA·TTC Trinucleotide Repeats Associated with Friedreich's Ataxia.Molecules. 2019 May 31;24(11):2078. doi: 10.3390/molecules24112078. Molecules. 2019. PMID: 31159174 Free PMC article.
-
Trinucleotide repeat instability during double-strand break repair: from mechanisms to gene therapy.Curr Genet. 2019 Feb;65(1):17-28. doi: 10.1007/s00294-018-0865-1. Epub 2018 Jul 5. Curr Genet. 2019. PMID: 29974202 Review.
-
Partners in crime: Tbf1 and Vid22 promote expansions of long human telomeric repeats at an interstitial chromosome position in yeast.PNAS Nexus. 2022 Jun 8;1(3):pgac080. doi: 10.1093/pnasnexus/pgac080. eCollection 2022 Jul. PNAS Nexus. 2022. PMID: 35832866 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

