MicroRNA-148a deficiency promotes hepatic lipid metabolism and hepatocarcinogenesis in mice
- PMID: 28703810
- PMCID: PMC5550856
- DOI: 10.1038/cddis.2017.309
MicroRNA-148a deficiency promotes hepatic lipid metabolism and hepatocarcinogenesis in mice
Abstract
miRNAs are involved in many physiologic and disease processes by virtue of degrading specific mRNAs or inhibiting their translation. miR-148a has been implicated in the control of tumor growth and cholesterol and triglyceride homeostasis using in vitro or in vivo gene expression- and silencing-based approaches. Here miR-148a knockout (KO) mice were used to investigate the intrinsic role of miR-148a in liver physiology and hepatocarcinogenesis in mice. miR-148a downregulation was found to be correlated with poor clinical outcomes in hepatocellular carcinoma (HCC) patients. Under regular chow diet (RCD) or high fat diet (HFD), miR-148a deletion significantly accelerated DEN-induced hepatocarcinogenesis in mice. Mechanistically, miR-148a deletion promotes lipid metabolic disorders in mice. Moreover, restoration of miR-148a reversed these defects. Finally, miR-148a was found to directly inhibit several key regulators of hepatocarcinogenesis and lipid metabolism. These findings reveal crucial roles for miR-148a in the hepatic lipid metabolism and hepatocarcinogenesis. They further identify miR-148a as a potential therapeutic target for certain liver diseases, including cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
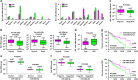

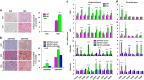
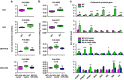

Similar articles
-
MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis.Int J Oncol. 2014 Jun;44(6):1915-22. doi: 10.3892/ijo.2014.2373. Epub 2014 Apr 8. Int J Oncol. 2014. PMID: 24714841
-
MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling.Oncogene. 2014 Jul 31;33(31):4069-76. doi: 10.1038/onc.2013.369. Epub 2013 Sep 9. Oncogene. 2014. PMID: 24013226
-
MiR-17-92 cluster promotes hepatocarcinogenesis.Carcinogenesis. 2015 Oct;36(10):1213-22. doi: 10.1093/carcin/bgv112. Epub 2015 Aug 1. Carcinogenesis. 2015. PMID: 26233958 Free PMC article.
-
The Role of Mir-148a in Cancer.J Cancer. 2016 Jun 21;7(10):1233-41. doi: 10.7150/jca.14616. eCollection 2016. J Cancer. 2016. PMID: 27390598 Free PMC article. Review.
-
Key regulatory miRNAs in lipid homeostasis: Implications for cardiometabolic diseases and development of novel therapeutics.Drug Discov Today. 2022 Aug;27(8):2170-2180. doi: 10.1016/j.drudis.2022.05.003. Epub 2022 May 10. Drug Discov Today. 2022. PMID: 35550438 Review.
Cited by
-
Evaluation of the performance of serum miRNAs as normalizers in microRNA studies focused on cardiovascular disease.J Thorac Dis. 2018 May;10(5):2599-2607. doi: 10.21037/jtd.2018.04.128. J Thorac Dis. 2018. PMID: 29997921 Free PMC article.
-
Control of hepatic gluconeogenesis by Argonaute2.Mol Metab. 2018 Dec;18:15-24. doi: 10.1016/j.molmet.2018.10.003. Epub 2018 Oct 9. Mol Metab. 2018. PMID: 30348590 Free PMC article.
-
Lipid metabolism in cancer progression and therapeutic strategies.MedComm (2020). 2020 Dec 24;2(1):27-59. doi: 10.1002/mco2.27. eCollection 2021 Mar. MedComm (2020). 2020. PMID: 34766135 Free PMC article. Review.
-
Regulation of hepatic microRNAs in response to early stage Echinococcus multilocularis egg infection in C57BL/6 mice.PLoS Negl Trop Dis. 2020 May 22;14(5):e0007640. doi: 10.1371/journal.pntd.0007640. eCollection 2020 May. PLoS Negl Trop Dis. 2020. PMID: 32442168 Free PMC article.
-
Inter-Individual Variability in Acute Toxicity of R-Pulegone and R-Menthofuran in Human Liver Slices and Their Influence on miRNA Expression Changes in Comparison to Acetaminophen.Int J Mol Sci. 2018 Jun 19;19(6):1805. doi: 10.3390/ijms19061805. Int J Mol Sci. 2018. PMID: 29921785 Free PMC article.
References
-
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132: 9–14. - PubMed
-
- Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 2015; 15: 653–667. - PubMed
-
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002; 31: 339–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

