Environmental change drives accelerated adaptation through stimulated copy number variation
- PMID: 28654659
- PMCID: PMC5486974
- DOI: 10.1371/journal.pbio.2001333
Environmental change drives accelerated adaptation through stimulated copy number variation
Abstract
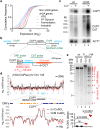
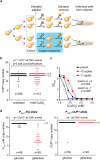
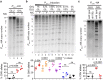
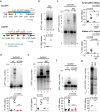
Copy number variation (CNV) is rife in eukaryotic genomes and has been implicated in many human disorders, particularly cancer, in which CNV promotes both tumorigenesis and chemotherapy resistance. CNVs are considered random mutations but often arise through replication defects; transcription can interfere with replication fork progression and stability, leading to increased mutation rates at highly transcribed loci. Here we investigate whether inducible promoters can stimulate CNV to yield reproducible, environment-specific genetic changes. We propose a general mechanism for environmentally-stimulated CNV and validate this mechanism for the emergence of copper resistance in budding yeast. By analysing a large cohort of individual cells, we directly demonstrate that CNV of the copper-resistance gene CUP1 is stimulated by environmental copper. CNV stimulation accelerates the formation of novel alleles conferring enhanced copper resistance, such that copper exposure actively drives adaptation to copper-rich environments. Furthermore, quantification of CNV in individual cells reveals remarkable allele selectivity in the rate at which specific environments stimulate CNV. We define the key mechanistic elements underlying this selectivity, demonstrating that CNV is regulated by both promoter activity and acetylation of histone H3 lysine 56 (H3K56ac) and that H3K56ac is required for CUP1 CNV and efficient copper adaptation. Stimulated CNV is not limited to high-copy CUP1 repeat arrays, as we find that H3K56ac also regulates CNV in 3 copy arrays of CUP1 or SFA1 genes. The impact of transcription on DNA damage is well understood, but our research reveals that this apparently problematic association forms a pathway by which mutations can be directed to particular loci in particular environments and furthermore that this mutagenic process can be regulated through histone acetylation. Stimulated CNV therefore represents an unanticipated and remarkably controllable pathway facilitating organismal adaptation to new environments.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: in addition to the funding described in the Financial Disclosure, JH declares that part of this work (although not the presented data), specifically the potential of histone acetyltransferase inhibition to prevent adaptation, forms part of a patent application.
Figures


Similar articles
-
The N-Terminal Tail of Histone H3 Regulates Copper Homeostasis in Saccharomyces cerevisiae.Mol Cell Biol. 2021 Jan 25;41(2):e00210-20. doi: 10.1128/MCB.00210-20. Print 2021 Jan 25. Mol Cell Biol. 2021. PMID: 33257505 Free PMC article.
-
Stimulation of adaptive gene amplification by origin firing under replication fork constraint.Nucleic Acids Res. 2022 Jan 25;50(2):915-936. doi: 10.1093/nar/gkab1257. Nucleic Acids Res. 2022. PMID: 35018465 Free PMC article.
-
Histone H2A and Spt10 cooperate to regulate induction and autoregulation of the CUP1 metallothionein.J Biol Chem. 2005 Jan 7;280(1):104-11. doi: 10.1074/jbc.M411437200. Epub 2004 Oct 21. J Biol Chem. 2005. PMID: 15501826
-
Production of metallothionein in copper- and cadmium-resistant strains of Saccharomyces cerevisiae.J Ind Microbiol. 1995 Feb;14(2):126-31. doi: 10.1007/BF01569894. J Ind Microbiol. 1995. PMID: 7766204 Review.
-
The molecular genetics of copper resistance in Saccharomyces cerevisiae--a paradigm for non-conventional yeasts.J Basic Microbiol. 1988;28(3):147-60. doi: 10.1002/jobm.3620280302. J Basic Microbiol. 1988. PMID: 3057171 Review.
Cited by
-
The distribution of beneficial mutational effects between two sister yeast species poorly explains natural outcomes of vineyard adaptation.bioRxiv [Preprint]. 2024 Jun 4:2024.06.03.597243. doi: 10.1101/2024.06.03.597243. bioRxiv. 2024. Update in: Genetics. 2024 Oct 07:iyae160. doi: 10.1093/genetics/iyae160 PMID: 38895255 Free PMC article. Updated. Preprint.
-
Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates.Curr Genet. 2019 Oct;65(5):1217-1228. doi: 10.1007/s00294-019-00976-w. Epub 2019 Apr 24. Curr Genet. 2019. PMID: 31020384 Free PMC article.
-
Population Structure, and Selection Signatures Underlying High-Altitude Adaptation Inferred From Genome-Wide Copy Number Variations in Chinese Indigenous Cattle.Front Genet. 2020 Feb 14;10:1404. doi: 10.3389/fgene.2019.01404. eCollection 2019. Front Genet. 2020. PMID: 32117428 Free PMC article.
-
Bridging Tumorigenesis and Therapy Resistance With a Non-Darwinian and Non-Lamarckian Mechanism of Adaptive Evolution.Front Oncol. 2021 Sep 10;11:732081. doi: 10.3389/fonc.2021.732081. eCollection 2021. Front Oncol. 2021. PMID: 34568068 Free PMC article.
-
Copy Number Variation and Allele Ratio Analysis in Candida albicans Using Whole Genome Sequencing Data.Methods Mol Biol. 2023;2658:105-125. doi: 10.1007/978-1-0716-3155-3_8. Methods Mol Biol. 2023. PMID: 37024698 Free PMC article.
References
-
- Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16(3):172–83. doi: 10.1038/nrg3871 . - DOI - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–8. doi: 10.1126/science.1098918 . - DOI - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–51. doi: 10.1038/ng1416 . - DOI - PubMed
-
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464(7289):713–20. doi: 10.1038/nature08979 ; - DOI - PMC - PubMed
-
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annual review of medicine. 2010;61:437–55. doi: 10.1146/annurev-med-100708-204735 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

