Single-molecule analysis of steroid receptor and cofactor action in living cells
- PMID: 28635963
- PMCID: PMC5482060
- DOI: 10.1038/ncomms15896
Single-molecule analysis of steroid receptor and cofactor action in living cells
Abstract
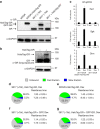
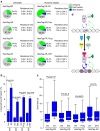
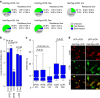
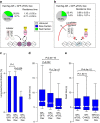
Population-based assays have been employed extensively to investigate the interactions of transcription factors (TFs) with chromatin and are often interpreted in terms of static and sequential binding. However, fluorescence microscopy techniques reveal a more dynamic binding behaviour of TFs in live cells. Here we analyse the strengths and limitations of in vivo single-molecule tracking and performed a comprehensive analysis on the intranuclear dwell times of four steroid receptors and a number of known cofactors. While the absolute residence times estimates can depend on imaging acquisition parameters due to sampling bias, our results indicate that only a small proportion of factors are specifically bound to chromatin at any given time. Interestingly, the glucocorticoid receptor and its cofactors affect each other's dwell times in an asymmetric manner. Overall, our data indicate transient rather than stable TF-cofactors chromatin interactions at response elements at the single-molecule level.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Glucocorticoid hormone-induced receptor localization to the chromatin fibers formed on injected DNA in Xenopus oocytes.Exp Cell Res. 2001 May 1;265(2):319-28. doi: 10.1006/excr.2001.5184. Exp Cell Res. 2001. PMID: 11302698
-
The glucocorticoid receptor and the orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II interact with and mutually affect each other's transcriptional activities: implications for intermediary metabolism.Mol Endocrinol. 2004 Apr;18(4):820-33. doi: 10.1210/me.2003-0341. Epub 2004 Jan 22. Mol Endocrinol. 2004. PMID: 14739255
-
Occupancy and composition of proteins bound to the AP-1 sites in the glucocorticoid receptor and c-jun promoters after glucocorticoid treatment and in different cell types.Recept Signal Transduct. 1996;6(3-4):179-93. Recept Signal Transduct. 1996. PMID: 9259052
-
The role of cofactors in sex steroid action.Best Pract Res Clin Endocrinol Metab. 2007 Sep;21(3):403-14. doi: 10.1016/j.beem.2007.07.002. Best Pract Res Clin Endocrinol Metab. 2007. PMID: 17875488 Review.
-
Interaction of the glucocorticoid receptor and the chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII): implications for the actions of glucocorticoids on glucose, lipoprotein, and xenobiotic metabolism.Ann N Y Acad Sci. 2004 Jun;1024:72-85. doi: 10.1196/annals.1321.006. Ann N Y Acad Sci. 2004. PMID: 15265774 Review.
Cited by
-
Inferring quantity and qualities of superimposed reaction rates from single molecule survival time distributions.Sci Rep. 2020 Feb 4;10(1):1758. doi: 10.1038/s41598-020-58634-y. Sci Rep. 2020. PMID: 32019978 Free PMC article.
-
Glucocorticoid-induced phosphorylation by CDK9 modulates the coactivator functions of transcriptional cofactor GRIP1 in macrophages.Nat Commun. 2017 Nov 23;8(1):1739. doi: 10.1038/s41467-017-01569-2. Nat Commun. 2017. PMID: 29170386 Free PMC article.
-
Epigenetic mechanisms in breast cancer therapy and resistance.Nat Commun. 2021 Mar 19;12(1):1786. doi: 10.1038/s41467-021-22024-3. Nat Commun. 2021. PMID: 33741974 Free PMC article. Review.
-
Compartmentalization of androgen receptors at endogenous genes in living cells.Nucleic Acids Res. 2023 Nov 10;51(20):10992-11009. doi: 10.1093/nar/gkad803. Nucleic Acids Res. 2023. PMID: 37791849 Free PMC article.
-
Transcriptional kinetic synergy: A complex landscape revealed by integrating modeling and synthetic biology.Cell Syst. 2023 Apr 19;14(4):324-339.e7. doi: 10.1016/j.cels.2023.02.003. Cell Syst. 2023. PMID: 37080164 Free PMC article.
References
-
- Mueller F., Stasevich T. J., Mazza D. & McNally J. G. Quantifying transcription factor kinetics: at work or at play? Crit. Rev. Biochem. Mol. Biol. 48, 492–514 (2013). - PubMed
-
- Thanos D. & Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100 (1995). - PubMed
-
- McNally J. G., Mueller W. G., Walker D., Wolford R. G. & Hager G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

