Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells
- PMID: 28628034
- PMCID: PMC5490740
- DOI: 10.1172/JCI93289
Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells
Abstract
HIV-1 causes a chronic, incurable disease due to its persistence in CD4+ T cells that contain replication-competent provirus, but exhibit little or no active viral gene expression and effectively resist combination antiretroviral therapy (cART). These latently infected T cells represent an extremely small proportion of all circulating CD4+ T cells but possess a remarkable long-term stability and typically persist throughout life, for reasons that are not fully understood. Here we performed massive single-genome, near-full-length next-generation sequencing of HIV-1 DNA derived from unfractionated peripheral blood mononuclear cells, ex vivo-isolated CD4+ T cells, and subsets of functionally polarized memory CD4+ T cells. This approach identified multiple sets of independent, near-full-length proviral sequences from cART-treated individuals that were completely identical, consistent with clonal expansion of CD4+ T cells harboring intact HIV-1. Intact, near-full-genome HIV-1 DNA sequences that were derived from such clonally expanded CD4+ T cells constituted 62% of all analyzed genome-intact sequences in memory CD4 T cells, were preferentially observed in Th1-polarized cells, were longitudinally detected over a duration of up to 5 years, and were fully replication- and infection-competent. Together, these data suggest that clonal proliferation of Th1-polarized CD4+ T cells encoding for intact HIV-1 represents a driving force for stabilizing the pool of latently infected CD4+ T cells.
Conflict of interest statement
Figures
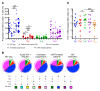
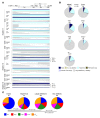
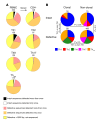
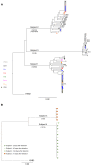
Comment in
- HIV persistence: clonal expansion of cells in the latent reservoir
Similar articles
-
Th1/17 Polarization of CD4 T Cells Supports HIV-1 Persistence during Antiretroviral Therapy.J Virol. 2015 Nov;89(22):11284-93. doi: 10.1128/JVI.01595-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339043 Free PMC article.
-
CD161+ CD4+ T Cells Harbor Clonally Expanded Replication-Competent HIV-1 in Antiretroviral Therapy-Suppressed Individuals.mBio. 2019 Oct 8;10(5):e02121-19. doi: 10.1128/mBio.02121-19. mBio. 2019. PMID: 31594817 Free PMC article.
-
HIV persistence: clonal expansion of cells in the latent reservoir.J Clin Invest. 2017 Jun 30;127(7):2536-2538. doi: 10.1172/JCI95329. Epub 2017 Jun 19. J Clin Invest. 2017. PMID: 28628041 Free PMC article.
-
Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy.Annu Rev Immunol. 2000;18:665-708. doi: 10.1146/annurev.immunol.18.1.665. Annu Rev Immunol. 2000. PMID: 10837072 Review.
-
The role of HIV integration in viral persistence: no more whistling past the proviral graveyard.J Clin Invest. 2016 Feb;126(2):438-47. doi: 10.1172/JCI80564. Epub 2016 Feb 1. J Clin Invest. 2016. PMID: 26829624 Free PMC article. Review.
Cited by
-
New Assay Reveals Vast Excess of Defective over Intact HIV-1 Transcripts in Antiretroviral Therapy-Suppressed Individuals.J Virol. 2022 Dec 21;96(24):e0160522. doi: 10.1128/jvi.01605-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448806 Free PMC article.
-
HIV Productively Infects Highly Differentiated and Exhausted CD4+ T Cells During AIDS.Pathog Immun. 2024 Feb 22;8(2):92-114. doi: 10.20411/pai.v8i2.638. eCollection 2023. Pathog Immun. 2024. PMID: 38420260 Free PMC article.
-
Tyrosine Kinase Inhibition: a New Perspective in the Fight against HIV.Curr HIV/AIDS Rep. 2019 Oct;16(5):414-422. doi: 10.1007/s11904-019-00462-5. Curr HIV/AIDS Rep. 2019. PMID: 31506864 Free PMC article. Review.
-
Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir.Proc Natl Acad Sci U S A. 2018 Aug 14;115(33):E7795-E7804. doi: 10.1073/pnas.1803468115. Epub 2018 Jul 30. Proc Natl Acad Sci U S A. 2018. PMID: 30061382 Free PMC article.
-
Immune correlates of HIV-1 reservoir cell decline in early-treated infants.Cell Rep. 2022 Jul 19;40(3):111126. doi: 10.1016/j.celrep.2022.111126. Cell Rep. 2022. PMID: 35858580 Free PMC article.
References
MeSH terms
Grants and funding
- R21 AI106468/AI/NIAID NIH HHS/United States
- R01 AI078799/AI/NIAID NIH HHS/United States
- R01 HL134539/HL/NHLBI NIH HHS/United States
- U01 AI114235/AI/NIAID NIH HHS/United States
- R01 AI098487/AI/NIAID NIH HHS/United States
- R37 AI067073/AI/NIAID NIH HHS/United States
- U01 AI117841/AI/NIAID NIH HHS/United States
- R33 AI116228/AI/NIAID NIH HHS/United States
- R21 AI124776/AI/NIAID NIH HHS/United States
- R01 AI120008/AI/NIAID NIH HHS/United States
- R21 AI116228/AI/NIAID NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

