Positively Selected Sites at HCMV gB Furin Processing Region and Their Effects in Cleavage Efficiency
- PMID: 28588572
- PMCID: PMC5441137
- DOI: 10.3389/fmicb.2017.00934
Positively Selected Sites at HCMV gB Furin Processing Region and Their Effects in Cleavage Efficiency
Abstract
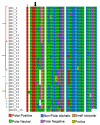
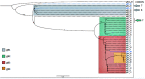
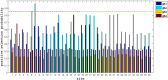
Human cytomegalovirus is a ubiquitous infectious agent that affects mainly immunosuppressed, fetuses, and newborns. The virus has several polymorphic regions, in particular in the envelope glycoproteins. The UL55 gene encodes the glycoprotein B that has a variable region, containing a furin cleavage site and according to the variability different genotypes are characterized. Here we investigated variability and existence of selective pressure on the UL55 variable region containing the furin cleavage site in 213 clinical sequences from patients worldwide. We showed the occurrence of positive selective pressure on gB codons 461 and 462, near the furin cleavage site. Cleavage analysis of synthesized peptides demonstrated that most mutations confer better cleavage by furin, suggesting that evolution is acting in order to increase the efficiency cleavage and supporting the hypothesis that gB processing is important in the host. We also demonstrated that peptides containing sequences, that characterize genotypes gB2 and 3, are differentially cleaved by furin. Our data demonstrate for the first time that variability in the cleavage site is related to degree of gB processing by furin.
Keywords: furin cleavage; gB genotypes; glycoprotein B; human cytomegalovirus; selective pressure.
Figures

Similar articles
-
Engineering glycoprotein B of bovine herpesvirus 1 to function as transporter for secreted proteins: a new protein expression approach.J Virol. 2005 Jan;79(2):791-9. doi: 10.1128/JVI.79.2.791-799.2005. J Virol. 2005. PMID: 15613307 Free PMC article.
-
Glycoprotein B of equine herpesvirus type 1 has two recognition sites for subtilisin-like proteases that are cleaved by furin.J Gen Virol. 2016 May;97(5):1218-1228. doi: 10.1099/jgv.0.000418. Epub 2016 Feb 3. J Gen Virol. 2016. PMID: 26843465
-
Proteolytic processing of human cytomegalovirus glycoprotein B is dispensable for viral growth in culture.J Virol. 2002 Feb;76(3):1252-64. doi: 10.1128/jvi.76.3.1252-1264.2002. J Virol. 2002. PMID: 11773401 Free PMC article.
-
[Determination of cytomegalovirus glycoprotein B genotypes in different geographical regions and different patient groups in Turkey].Mikrobiyol Bul. 2016 Jan;50(1):53-62. doi: 10.5578/mb.10880. Mikrobiyol Bul. 2016. PMID: 27058329 Turkish.
-
Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones.Biochimie. 1994;76(3-4):217-25. doi: 10.1016/0300-9084(94)90149-x. Biochimie. 1994. PMID: 7819326 Review.
Cited by
-
Evolution and Genetic Diversity of Primate Cytomegaloviruses.Microorganisms. 2020 Apr 25;8(5):624. doi: 10.3390/microorganisms8050624. Microorganisms. 2020. PMID: 32344906 Free PMC article. Review.
-
HCMV Envelope Glycoprotein Diversity Demystified.Front Microbiol. 2019 May 15;10:1005. doi: 10.3389/fmicb.2019.01005. eCollection 2019. Front Microbiol. 2019. PMID: 31156572 Free PMC article.
-
SARS-CoV-2 Spike Protein Is Capable of Inducing Cell-Cell Fusions Independent from Its Receptor ACE2 and This Activity Can Be Impaired by Furin Inhibitors or a Subset of Monoclonal Antibodies.Viruses. 2023 Jul 4;15(7):1500. doi: 10.3390/v15071500. Viruses. 2023. PMID: 37515187 Free PMC article.
-
Whole-Genome Sequence Analysis of Pseudorabies Virus Clinical Isolates from Pigs in China between 2012 and 2017 in China.Viruses. 2021 Jul 8;13(7):1322. doi: 10.3390/v13071322. Viruses. 2021. PMID: 34372529 Free PMC article.
-
Past and ongoing adaptation of human cytomegalovirus to its host.PLoS Pathog. 2020 May 8;16(5):e1008476. doi: 10.1371/journal.ppat.1008476. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32384127 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

