Signalome-wide RNAi screen identifies GBA1 as a positive mediator of autophagic cell death
- PMID: 28574511
- PMCID: PMC5520177
- DOI: 10.1038/cdd.2017.80
Signalome-wide RNAi screen identifies GBA1 as a positive mediator of autophagic cell death
Abstract
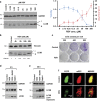
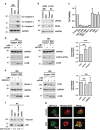
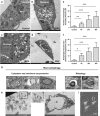
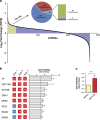
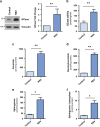
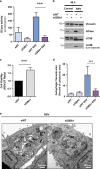
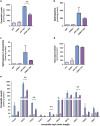
Activating alternative cell death pathways, including autophagic cell death, is a promising direction to overcome the apoptosis resistance observed in various cancers. Yet, whether autophagy acts as a death mechanism by over consumption of intracellular components is still controversial and remains undefined at the ultrastructural and the mechanistic levels. Here we identified conditions under which resveratrol-treated A549 lung cancer cells die by a mechanism that fulfills the previous definition of autophagic cell death. The cells displayed a strong and sustained induction of autophagic flux, cell death was prevented by knocking down autophagic genes and death occurred in the absence of apoptotic or necroptotic pathway activation. Detailed ultrastructural characterization revealed additional critical events, including a continuous increase over time in the number of autophagic vacuoles, in particular autolysosomes, occupying most of the cytoplasm at terminal stages. This was followed by loss of organelles, disruption of intracellular membranes including the swelling of perinuclear space and, occasionally, a unique type of nuclear shedding. A signalome-wide shRNA-based viability screen was applied to identify positive mediators of this type of autophagic cell death. One top hit was GBA1, the Gaucher disease-associated gene, which encodes glucocerebrosidase, an enzyme that metabolizes glucosylceramide to ceramide and glucose. Interestingly, glucocerebrosidase expression levels and activity were elevated, concomitantly with increased intracellular ceramide levels, both of which correlated in time with the appearance of the unique death characteristics. Transfection with siGBA1 attenuated the increase in glucocerebrosidase activity and the intracellular ceramide levels. Most importantly, GBA1 knockdown prevented the strong increase in LC3 lipidation, and many of the ultrastructural changes characteristic of this type of autophagic cell death, including a significant decrease in cytoplasmic area occupied by autophagic vacuoles. Together, these findings highlight the critical role of GBA1 in mediating enhanced self-consumption of intracellular components and endomembranes, leading to autophagic cell death.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Death by over-eating: The Gaucher disease associated gene GBA1, identified in a screen for mediators of autophagic cell death, is necessary for developmental cell death in Drosophila midgut.Cell Cycle. 2017;16(21):2003-2010. doi: 10.1080/15384101.2017.1380134. Epub 2017 Oct 9. Cell Cycle. 2017. PMID: 28933588 Free PMC article. Review.
-
A Drosophila Model of Neuronopathic Gaucher Disease Demonstrates Lysosomal-Autophagic Defects and Altered mTOR Signalling and Is Functionally Rescued by Rapamycin.J Neurosci. 2016 Nov 16;36(46):11654-11670. doi: 10.1523/JNEUROSCI.4527-15.2016. J Neurosci. 2016. PMID: 27852774 Free PMC article.
-
Resveratrol-induced autophagy and apoptosis in cisplatin-resistant human oral cancer CAR cells: A key role of AMPK and Akt/mTOR signaling.Int J Oncol. 2017 Mar;50(3):873-882. doi: 10.3892/ijo.2017.3866. Epub 2017 Jan 30. Int J Oncol. 2017. PMID: 28197628
-
Resveratrol analogue 3,4,4'-trihydroxy-trans-stilbene induces apoptosis and autophagy in human non-small-cell lung cancer cells in vitro.Acta Pharmacol Sin. 2015 Oct;36(10):1256-65. doi: 10.1038/aps.2015.46. Epub 2015 Jul 20. Acta Pharmacol Sin. 2015. PMID: 26190500 Free PMC article.
-
Cell death and autophagy: cytokines, drugs, and nutritional factors.Toxicology. 2008 Dec 30;254(3):147-57. doi: 10.1016/j.tox.2008.07.048. Epub 2008 Jul 23. Toxicology. 2008. PMID: 18694801 Review.
Cited by
-
Chemotherapy Resistance: Role of Mitochondrial and Autophagic Components.Cancers (Basel). 2022 Mar 12;14(6):1462. doi: 10.3390/cancers14061462. Cancers (Basel). 2022. PMID: 35326612 Free PMC article. Review.
-
Glycolipid Metabolite β-Glucosylceramide Is a Neutrophil Extracellular Trap-Inducing Ligand of Mincle Released during Bacterial Infection and Inflammation.J Immunol. 2022 Jul 15;209(2):391-400. doi: 10.4049/jimmunol.2100855. Epub 2022 Jun 29. J Immunol. 2022. PMID: 35768151 Free PMC article.
-
Distinct Signaling Pathways for Autophagy-Driven Cell Death and Survival in Adult Hippocampal Neural Stem Cells.Int J Mol Sci. 2023 May 5;24(9):8289. doi: 10.3390/ijms24098289. Int J Mol Sci. 2023. PMID: 37175992 Free PMC article.
-
Biological Functions of Autophagy Genes: A Disease Perspective.Cell. 2019 Jan 10;176(1-2):11-42. doi: 10.1016/j.cell.2018.09.048. Cell. 2019. PMID: 30633901 Free PMC article. Review.
-
Cell Survival and Cell Death at the Intersection of Autophagy and Apoptosis: Implications for Current and Future Cancer Therapeutics.ACS Pharmacol Transl Sci. 2021 Nov 3;4(6):1728-1746. doi: 10.1021/acsptsci.1c00130. eCollection 2021 Dec 10. ACS Pharmacol Transl Sci. 2021. PMID: 34927007 Free PMC article. Review.
References
-
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147: 728–741. - PubMed
-
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem 2005; 280: 20722–20729. - PubMed
-
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009; 137: 721–735. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

