The Role of Somatic L1 Retrotransposition in Human Cancers
- PMID: 28561751
- PMCID: PMC5490808
- DOI: 10.3390/v9060131
The Role of Somatic L1 Retrotransposition in Human Cancers
Abstract
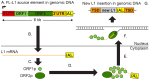
The human LINE-1 (or L1) element is a non-LTR retrotransposon that is mobilized through an RNA intermediate by an L1-encoded reverse transcriptase and other L1-encoded proteins. L1 elements remain actively mobile today and continue to mutagenize human genomes. Importantly, when new insertions disrupt gene function, they can cause diseases. Historically, L1s were thought to be active in the germline but silenced in adult somatic tissues. However, recent studies now show that L1 is active in at least some somatic tissues, including epithelial cancers. In this review, we provide an overview of these recent developments, and examine evidence that somatic L1 retrotransposition can initiate and drive tumorigenesis in humans. Recent studies have: (i) cataloged somatic L1 activity in many epithelial tumor types; (ii) identified specific full-length L1 source elements that give rise to somatic L1 insertions; and (iii) determined that L1 promoter hypomethylation likely plays an early role in the derepression of L1s in somatic tissues. A central challenge moving forward is to determine the extent to which L1 driver mutations can promote tumor initiation, evolution, and metastasis in humans.
Keywords: LINE-1, L1; cancer genomics; retrotransposon; somatic retrotransposition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
L1 Mosaicism in Mammals: Extent, Effects, and Evolution.Trends Genet. 2017 Nov;33(11):802-816. doi: 10.1016/j.tig.2017.07.004. Epub 2017 Aug 7. Trends Genet. 2017. PMID: 28797643 Review.
-
Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers.Genome Res. 2018 Aug;28(8):1136-1146. doi: 10.1101/gr.231837.117. Epub 2018 Jul 3. Genome Res. 2018. PMID: 29970450 Free PMC article.
-
Widespread somatic L1 retrotransposition in normal colorectal epithelium.Nature. 2023 May;617(7961):540-547. doi: 10.1038/s41586-023-06046-z. Epub 2023 May 10. Nature. 2023. PMID: 37165195 Free PMC article.
-
A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer.Genome Res. 2016 Jun;26(6):745-55. doi: 10.1101/gr.201814.115. Epub 2016 May 10. Genome Res. 2016. PMID: 27197217 Free PMC article.
-
L1 retrotransposons and somatic mosaicism in the brain.Annu Rev Genet. 2014;48:1-27. doi: 10.1146/annurev-genet-120213-092412. Epub 2014 Jul 14. Annu Rev Genet. 2014. PMID: 25036377 Review.
Cited by
-
Distinct contributions of DNA methylation and histone acetylation to the genomic occupancy of transcription factors.Genome Res. 2020 Oct;30(10):1393-1406. doi: 10.1101/gr.257576.119. Epub 2020 Sep 22. Genome Res. 2020. PMID: 32963030 Free PMC article.
-
L1 chimeric transcripts are expressed in healthy brain and their deregulation in glioma follows that of their host locus.Hum Mol Genet. 2022 Aug 17;31(15):2606-2622. doi: 10.1093/hmg/ddac056. Hum Mol Genet. 2022. PMID: 35298627 Free PMC article.
-
HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity.Front Microbiol. 2018 Mar 14;9:462. doi: 10.3389/fmicb.2018.00462. eCollection 2018. Front Microbiol. 2018. PMID: 29593697 Free PMC article. Review.
-
Precise characterization of somatic complex structural variations from tumor/control paired long-read sequencing data with nanomonsv.Nucleic Acids Res. 2023 Aug 11;51(14):e74. doi: 10.1093/nar/gkad526. Nucleic Acids Res. 2023. PMID: 37336583 Free PMC article.
-
Towards targeting transposable elements for cancer therapy.Nat Rev Cancer. 2024 Feb;24(2):123-140. doi: 10.1038/s41568-023-00653-8. Epub 2024 Jan 16. Nat Rev Cancer. 2024. PMID: 38228901 Review.
References
-
- Eickbush T.H., Malik H.S. Mobile DNA II. ASM Press; Washington, DC, USA: 2002. Origins and Evolution of Retrotransposons.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

