WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy
- PMID: 28561066
- PMCID: PMC5460038
- DOI: 10.1038/ncomms15637
WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy
Abstract
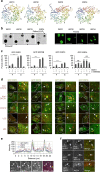
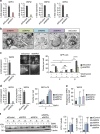
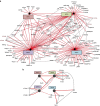
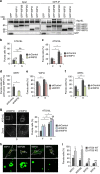
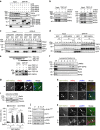
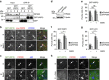
Autophagy is controlled by AMPK and mTOR, both of which associate with ULK1 and control the production of phosphatidylinositol 3-phosphate (PtdIns3P), a prerequisite for autophagosome formation. Here we report that WIPI3 and WIPI4 scaffold the signal control of autophagy upstream of PtdIns3P production and have a role in the PtdIns3P effector function of WIPI1-WIPI2 at nascent autophagosomes. In response to LKB1-mediated AMPK stimulation, WIPI4-ATG2 is released from a WIPI4-ATG2/AMPK-ULK1 complex and translocates to nascent autophagosomes, controlling their size, to which WIPI3, in complex with FIP200, also contributes. Upstream, WIPI3 associates with AMPK-activated TSC complex at lysosomes, regulating mTOR. Our WIPI interactome analysis reveals the scaffold functions of WIPI proteins interconnecting autophagy signal control and autophagosome formation. Our functional kinase screen uncovers a novel regulatory link between LKB1-mediated AMPK stimulation that produces a direct signal via WIPI4, and we show that the AMPK-related kinases NUAK2 and BRSK2 regulate autophagy through WIPI4.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
WIPI β-propellers function as scaffolds for STK11/LKB1-AMPK and AMPK-related kinase signaling in autophagy.Autophagy. 2018;14(6):1082-1083. doi: 10.1080/15548627.2017.1382784. Epub 2018 Feb 3. Autophagy. 2018. PMID: 28976799 Free PMC article.
-
WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome.J Cell Sci. 2015 Jan 15;128(2):207-17. doi: 10.1242/jcs.146258. J Cell Sci. 2015. PMID: 25568150 Review.
-
Dynamic association of the ULK1 complex with omegasomes during autophagy induction.J Cell Sci. 2013 Nov 15;126(Pt 22):5224-38. doi: 10.1242/jcs.132415. Epub 2013 Sep 6. J Cell Sci. 2013. PMID: 24013547
-
ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis.Autophagy. 2017 Aug 3;13(8):1386-1403. doi: 10.1080/15548627.2017.1332565. Epub 2017 Jul 11. Autophagy. 2017. PMID: 28696138 Free PMC article.
-
Human WIPI β-propeller function in autophagy and neurodegeneration.FEBS Lett. 2024 Jan;598(1):127-139. doi: 10.1002/1873-3468.14782. Epub 2023 Dec 14. FEBS Lett. 2024. PMID: 38058212 Review.
Cited by
-
WIPI1 promotes fission of endosomal transport carriers and formation of autophagosomes through distinct mechanisms.Autophagy. 2021 Nov;17(11):3644-3670. doi: 10.1080/15548627.2021.1886830. Epub 2021 Mar 8. Autophagy. 2021. PMID: 33685363 Free PMC article.
-
Functional evidence for a de novo mutation in WDR45 leading to BPAN in a Chinese girl.Mol Genet Genomic Med. 2019 Sep;7(9):e858. doi: 10.1002/mgg3.858. Epub 2019 Jul 22. Mol Genet Genomic Med. 2019. PMID: 31332960 Free PMC article.
-
El-Hattab-Alkuraya syndrome caused by biallelic WDR45B pathogenic variants: Further delineation of the phenotype and genotype.Clin Genet. 2022 May;101(5-6):530-540. doi: 10.1111/cge.14132. Epub 2022 Apr 12. Clin Genet. 2022. PMID: 35322404 Free PMC article.
-
LKB1 suppresses glioma cell invasion via NF-κB/Snail signaling repression.Onco Targets Ther. 2019 Apr 2;12:2451-2463. doi: 10.2147/OTT.S193736. eCollection 2019. Onco Targets Ther. 2019. PMID: 31040689 Free PMC article.
-
Autophagy in Age-Associated Neurodegeneration.Cells. 2018 May 5;7(5):37. doi: 10.3390/cells7050037. Cells. 2018. PMID: 29734735 Free PMC article. Review.
References
-
- Lamb C. A., Yoshimori T. & Tooze S. A. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 (2013). - PubMed
-
- Stolz A., Ernst A. & Dikic I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16, 495–501 (2014). - PubMed
-
- Rubinsztein D. C., Marino G. & Kroemer G. Autophagy and aging. Cell 146, 682–695 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

