A Naturally Occurring Recombinant Enterovirus Expresses a Torovirus Deubiquitinase
- PMID: 28490584
- PMCID: PMC5487566
- DOI: 10.1128/JVI.00450-17
A Naturally Occurring Recombinant Enterovirus Expresses a Torovirus Deubiquitinase
Abstract
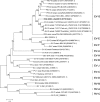
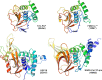
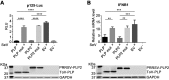
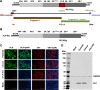
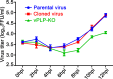
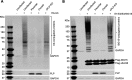
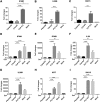
Enteroviruses (EVs) are implicated in a wide range of diseases in humans and animals. In this study, a novel enterovirus (enterovirus species G [EVG]) (EVG 08/NC_USA/2015) was isolated from a diagnostic sample from a neonatal pig diarrhea case and identified by using metagenomics and complete genome sequencing. The viral genome shares 75.4% nucleotide identity with a prototypic EVG strain (PEV9 UKG/410/73). Remarkably, a 582-nucleotide insertion, flanked by 3Cpro cleavage sites at the 5' and 3' ends, was found in the 2C/3A junction region of the viral genome. This insertion encodes a predicted protease with 54 to 68% amino acid identity to torovirus (ToV) papain-like protease (PLP) (ToV-PLP). Structural homology modeling predicts that this protease adopts a fold and a catalytic site characteristic of minimal PLP catalytic domains. This structure is similar to those of core catalytic domains of the foot-and-mouth disease virus leader protease and coronavirus PLPs, which act as deubiquitinating and deISGylating (interferon [IFN]-stimulated gene 15 [ISG15]-removing) enzymes on host cell substrates. Importantly, the recombinant ToV-PLP protein derived from this novel enterovirus also showed strong deubiquitination and deISGylation activities and demonstrated the ability to suppress IFN-β expression. Using reverse genetics, we generated a ToV-PLP knockout recombinant virus. Compared to the wild-type virus, the ToV-PLP knockout mutant virus showed impaired growth and induced higher expression levels of innate immune genes in infected cells. These results suggest that ToV-PLP functions as an innate immune antagonist; enterovirus G may therefore gain fitness through the acquisition of ToV-PLP from a recombination event.IMPORTANCE Enteroviruses comprise a highly diversified group of viruses. Genetic recombination has been considered a driving force for viral evolution; however, recombination between viruses from two different orders is a rare event. In this study, we identified a special case of cross-order recombination between enterovirus G (order Picornavirales) and torovirus (order Nidovirales). This naturally occurring recombination event may have broad implications for other picornaviral and/or nidoviral species. Importantly, we demonstrated that the exogenous ToV-PLP gene that was inserted into the EVG genome encodes a deubiquitinase/deISGylase and potentially suppresses host cellular innate immune responses. Our results provide insights into how a gain of function through genetic recombination, in particular cross-order recombination, may improve the ability of a virus to evade host immunity.
Keywords: deubiquitinase; enterovirus G; genetic recombination; papain-like protease; torovirus.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
First detection of novel enterovirus G recombining a torovirus papain-like protease gene associated with diarrhoea in swine in South Korea.Transbound Emerg Dis. 2019 Mar;66(2):1023-1028. doi: 10.1111/tbed.13073. Epub 2018 Dec 1. Transbound Emerg Dis. 2019. PMID: 30431236 Free PMC article.
-
Full-length and defective enterovirus G genomes with distinct torovirus protease insertions are highly prevalent on a Chinese pig farm.Arch Virol. 2018 Sep;163(9):2471-2476. doi: 10.1007/s00705-018-3875-x. Epub 2018 May 21. Arch Virol. 2018. PMID: 29786119
-
A novel defective recombinant porcine enterovirus G virus carrying a porcine torovirus papain-like cysteine protease gene and a putative anti-apoptosis gene in place of viral structural protein genes.Infect Genet Evol. 2019 Nov;75:103975. doi: 10.1016/j.meegid.2019.103975. Epub 2019 Jul 22. Infect Genet Evol. 2019. PMID: 31344488 Free PMC article.
-
Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities.Virus Res. 2014 Dec 19;194:184-90. doi: 10.1016/j.virusres.2014.01.025. Epub 2014 Feb 7. Virus Res. 2014. PMID: 24512893 Free PMC article. Review.
-
Role of recombination in evolution of enteroviruses.Rev Med Virol. 2005 May-Jun;15(3):157-67. doi: 10.1002/rmv.457. Rev Med Virol. 2005. PMID: 15578739 Review.
Cited by
-
Structure and Function of Viral Deubiquitinating Enzymes.J Mol Biol. 2017 Nov 10;429(22):3441-3470. doi: 10.1016/j.jmb.2017.06.010. Epub 2017 Jun 16. J Mol Biol. 2017. PMID: 28625850 Free PMC article. Review.
-
Recent Progress in Torovirus Molecular Biology.Viruses. 2021 Mar 8;13(3):435. doi: 10.3390/v13030435. Viruses. 2021. PMID: 33800523 Free PMC article. Review.
-
First isolation, identification, and pathogenicity evaluation of an EV-G6 strain in China.Front Vet Sci. 2024 Jul 24;11:1431180. doi: 10.3389/fvets.2024.1431180. eCollection 2024. Front Vet Sci. 2024. PMID: 39113722 Free PMC article.
-
Porcine Torovirus (PToV)-A Brief Review of Etiology, Diagnostic Assays and Current Epidemiology.Front Vet Sci. 2019 Apr 18;6:120. doi: 10.3389/fvets.2019.00120. eCollection 2019. Front Vet Sci. 2019. PMID: 31058174 Free PMC article. Review.
-
Characterization and Identification of a Novel Torovirus Associated With Recombinant Bovine Torovirus From Tibetan Antelope in Qinghai-Tibet Plateau of China.Front Microbiol. 2021 Sep 6;12:737753. doi: 10.3389/fmicb.2021.737753. eCollection 2021. Front Microbiol. 2021. PMID: 34552576 Free PMC article.
References
-
- Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. 2012. Virus taxonomy: classification and nomenclature of viruses, p 855–880. In King AMQ, ELefkowitz E, Adams MJ, Carstens EB (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

