NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain
- PMID: 28465465
- PMCID: PMC5460996
- DOI: 10.1084/jem.20160933
NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain
Abstract
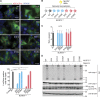
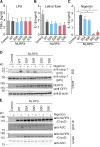
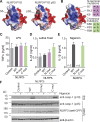
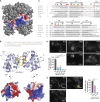
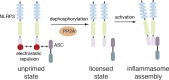
NLRP3 is a cytosolic pattern recognition receptor that senses microbes and endogenous danger signals. Upon activation, NLRP3 forms an inflammasome with the adapter ASC, resulting in caspase-1 activation, release of proinflammatory cytokines and cell death. How NLRP3 activation is regulated by transcriptional and posttranslational mechanisms to prevent aberrant activation remains incompletely understood. Here, we identify three conserved phosphorylation sites in NLRP3 and demonstrate that NLRP3 activation is controlled by phosphorylation of its pyrin domain (PYD). Phosphomimetic residues in NLRP3 PYD abrogate inflammasome activation and structural modeling indicates that phosphorylation of the PYD regulates charge-charge interaction between two PYDs that are essential for NLRP3 activation. Phosphatase 2A (PP2A) inhibition or knock-down drastically reduces NLRP3 activation, showing that PP2A can license inflammasome assembly via dephosphorylating NLRP3 PYD. These results propose that the balance between kinases and phosphatases acting on the NLRP3 PYD is critical for NLRP3 activation.
© 2017 Stutz et al.
Figures

Similar articles
-
The mechanism of NLRP3 inflammasome initiation: Trimerization but not dimerization of the NLRP3 pyrin domain induces robust activation of IL-1β.Biochem Biophys Res Commun. 2017 Feb 5;483(2):823-828. doi: 10.1016/j.bbrc.2017.01.008. Epub 2017 Jan 5. Biochem Biophys Res Commun. 2017. PMID: 28065854
-
Regulation of NLRP3 Inflammasome by Phosphorylation.Front Immunol. 2018 Oct 8;9:2305. doi: 10.3389/fimmu.2018.02305. eCollection 2018. Front Immunol. 2018. PMID: 30349539 Free PMC article. Review.
-
POP1 might be recruiting its type-Ia interface for NLRP3-mediated PYD-PYD interaction: Insights from MD simulation.J Mol Recognit. 2017 Sep;30(9). doi: 10.1002/jmr.2632. Epub 2017 Apr 3. J Mol Recognit. 2017. PMID: 28370480
-
Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein.J Biol Chem. 2012 Dec 7;287(50):41732-43. doi: 10.1074/jbc.M112.381228. Epub 2012 Oct 12. J Biol Chem. 2012. PMID: 23066025 Free PMC article.
-
Inhibiting the inflammasome: one domain at a time.Immunol Rev. 2015 May;265(1):205-16. doi: 10.1111/imr.12290. Immunol Rev. 2015. PMID: 25879295 Free PMC article. Review.
Cited by
-
The NLRP3 inflammasome: activation and regulation.Trends Biochem Sci. 2023 Apr;48(4):331-344. doi: 10.1016/j.tibs.2022.10.002. Epub 2022 Nov 4. Trends Biochem Sci. 2023. PMID: 36336552 Free PMC article. Review.
-
The Role of NLRP3 Inflammasome in the Pathogenesis of Traumatic Brain Injury.Int J Mol Sci. 2020 Aug 27;21(17):6204. doi: 10.3390/ijms21176204. Int J Mol Sci. 2020. PMID: 32867310 Free PMC article. Review.
-
Priming Is Dispensable for NLRP3 Inflammasome Activation in Human Monocytes In Vitro.Front Immunol. 2020 Sep 30;11:565924. doi: 10.3389/fimmu.2020.565924. eCollection 2020. Front Immunol. 2020. PMID: 33101286 Free PMC article.
-
NLRP3 lacking the leucine-rich repeat domain can be fully activated via the canonical inflammasome pathway.Nat Commun. 2018 Dec 5;9(1):5182. doi: 10.1038/s41467-018-07573-4. Nat Commun. 2018. PMID: 30518920 Free PMC article.
-
Crosstalk Between the NLRP3 Inflammasome/ASC Speck and Amyloid Protein Aggregates Drives Disease Progression in Alzheimer's and Parkinson's Disease.Front Mol Neurosci. 2022 Feb 3;15:805169. doi: 10.3389/fnmol.2022.805169. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35185469 Free PMC article. Review.
References
-
- Bauernfeind F.G., Horvath G., Stutz A., Alnemri E.S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B.G., Fitzgerald K.A., et al. . 2009. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183:787–791. 10.4049/jimmunol.0901363 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

