rs10499194 polymorphism in the tumor necrosis factor-α inducible protein 3 (TNFAIP3) gene is associated with type-1 autoimmune hepatitis risk in Chinese Han population
- PMID: 28448618
- PMCID: PMC5407796
- DOI: 10.1371/journal.pone.0176471
rs10499194 polymorphism in the tumor necrosis factor-α inducible protein 3 (TNFAIP3) gene is associated with type-1 autoimmune hepatitis risk in Chinese Han population
Retraction in
-
Retraction: rs10499194 polymorphism in the tumor necrosis factor-α inducible protein 3 (TNFAIP3) gene is associated with type-1 autoimmune hepatitis risk in Chinese Han population.PLoS One. 2018 Mar 27;13(3):e0195181. doi: 10.1371/journal.pone.0195181. eCollection 2018. PLoS One. 2018. PMID: 29584776 Free PMC article. No abstract available.
Abstract
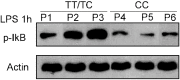
Previous studies have found that the polymorphisms of tumor necrosis factor-α induced protein 3 (TNFAIP3) were associated with several autoimmune diseases. However, the role of TNFAIP3 polymorphisms in type-1 autoimmune hepatitis (AIH-1) remained unclear. The present study aimed to clarify the association of TNFAIP3 polymorphisms with AIH-1 risk in a Chinese Han population. The TaqMan SNP genotyping assay was used to determine the distribution of TNFAIP3 polymorphisms in 432 AIH-1 patients and 500 healthy controls. The association of TNFAIP3 polymorphisms and clinical characteristic was further evaluated. Five TNFAIP3 polymorphisms (rs2230926, rs5029939, rs10499194, rs6920220, rs582757) were analyzed in the present study. No significant association could be observed between rs2230926, rs5029939, rs6920220, rs582757 and the susceptibility to AIH-1 in Chinese Han population. Compared with wild-type genotype CC at rs10499194, individuals carrying CT genotype had a significantly increased risk for developing AIH-1 (OR = 2.32, 95%CI 1.44-3.74). Under a dominant model, CT/TT carriers have a 140% increased risk of AIH-1 than CC carriers (OR = 2.40, 95%CI 1.50-3.87). The rs10499194 T allele was also found to be significantly associated with AIH-1 risk (OR = 2.41, 95%CI 1.51-3.82). In addition, higher serum ALT, AST levels and more common cirrhosis were observed in AIH-1 patients with T allele (CT/TT) than those with CC genotype. In conclusion, TNFAIP3 rs10499194 T allele and CT genotype were associated with an increased risk for AIH-1, suggesting rs10499194 polymorphism as a candidate of susceptibility locus to AIH-1.
Conflict of interest statement
Figures
Similar articles
-
Single nucleotide polymorphisms in TNFAIP3 were associated with the risks of rheumatoid arthritis in northern Chinese Han population.BMC Med Genet. 2014 May 15;15:56. doi: 10.1186/1471-2350-15-56. BMC Med Genet. 2014. PMID: 24884566 Free PMC article.
-
TNFAIP3 genetic polymorphisms reduce ankylosing spondylitis risk in Eastern Chinese Han population.Sci Rep. 2019 Jul 15;9(1):10209. doi: 10.1038/s41598-019-46647-1. Sci Rep. 2019. PMID: 31308453 Free PMC article.
-
Role of deleterious single nucleotide variants in the coding regions of TNFAIP3 for Japanese autoimmune hepatitis with cirrhosis.Sci Rep. 2019 May 28;9(1):7925. doi: 10.1038/s41598-019-44524-5. Sci Rep. 2019. PMID: 31138864 Free PMC article.
-
TNFAIP3 gene rs10499194, rs13207033 polymorphisms decrease the risk of rheumatoid arthritis.Oncotarget. 2016 Dec 13;7(50):82933-82942. doi: 10.18632/oncotarget.12638. Oncotarget. 2016. PMID: 27779104 Free PMC article. Review.
-
Autoimmune hepatitis, HLA and extended haplotypes.Autoimmun Rev. 2011 Feb;10(4):189-93. doi: 10.1016/j.autrev.2010.09.024. Epub 2010 Oct 7. Autoimmun Rev. 2011. PMID: 20933106 Review.
Cited by
-
Effects of Sepantronium Bromide (YM-155) on the Whole Transcriptome of MDA-MB-231 Cells: Highlight on Impaired ATR/ATM Fanconi Anemia DNA Damage Response.Cancer Genomics Proteomics. 2018 Jul-Aug;15(4):249-264. doi: 10.21873/cgp.20083. Cancer Genomics Proteomics. 2018. PMID: 29976630 Free PMC article.
-
Retraction: rs10499194 polymorphism in the tumor necrosis factor-α inducible protein 3 (TNFAIP3) gene is associated with type-1 autoimmune hepatitis risk in Chinese Han population.PLoS One. 2018 Mar 27;13(3):e0195181. doi: 10.1371/journal.pone.0195181. eCollection 2018. PLoS One. 2018. PMID: 29584776 Free PMC article. No abstract available.
-
Autoimmune Hepatitis-Immunologically Triggered Liver Pathogenesis-Diagnostic and Therapeutic Strategies.J Immunol Res. 2019 Nov 25;2019:9437043. doi: 10.1155/2019/9437043. eCollection 2019. J Immunol Res. 2019. PMID: 31886312 Free PMC article. Review.
-
A20/Tumor Necrosis Factor α-Induced Protein 3 in Immune Cells Controls Development of Autoinflammation and Autoimmunity: Lessons from Mouse Models.Front Immunol. 2018 Feb 21;9:104. doi: 10.3389/fimmu.2018.00104. eCollection 2018. Front Immunol. 2018. PMID: 29515565 Free PMC article. Review.
-
The progress of autoimmune hepatitis research and future challenges.Open Med (Wars). 2023 Oct 30;18(1):20230823. doi: 10.1515/med-2023-0823. eCollection 2023. Open Med (Wars). 2023. PMID: 38025543 Free PMC article. Review.
References
-
- Czaja AJ, Manns MP. Advances in the diagnosis, pathogenesis, and management of autoimmune hepatitis. Gastroenterology. 2010;139:58–72 e54. doi: 10.1053/j.gastro.2010.04.053 - DOI - PubMed
-
- Krawitt EL. Autoimmune hepatitis. N Engl J Med. 2006;354:54–66. doi: 10.1056/NEJMra050408 - DOI - PubMed
-
- Chen S, Zhao W, Tan W, Luo X, Dan Y, You Z, Kuang X, et al. Association of TBX21 promoter polymorphisms with type 1 autoimmune hepatitis in a Chinese population. Hum Immunol. 2011;72:69–73. doi: 10.1016/j.humimm.2010.10.019 - DOI - PubMed
-
- Migita K, Nakamura M, Abiru S, Jiuchi Y, Nagaoka S, Komori A, Hashimoto S, et al. Association of STAT4 polymorphisms with susceptibility to type-1 autoimmune hepatitis in the Japanese population. PLoS One. 2013;8:e71382 doi: 10.1371/journal.pone.0071382 - DOI - PMC - PubMed
-
- Longhi MS, Ma Y, Mieli-Vergani G, Vergani D. Aetiopathogenesis of autoimmune hepatitis. J Autoimmun. 2010;34:7–14. doi: 10.1016/j.jaut.2009.08.010 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources