A Novel Method to Evaluate Ribosomal Performance in Cell-Free Protein Synthesis Systems
- PMID: 28436469
- PMCID: PMC5402277
- DOI: 10.1038/srep46753
A Novel Method to Evaluate Ribosomal Performance in Cell-Free Protein Synthesis Systems
Abstract
Cell-free protein synthesis (CFPS) systems were designed to produce proteins with a minimal set of purified components, thus offering the possibility to follow translation as well as protein folding. In order to characterize the performance of the ribosomes in such a system, it is crucial to separately quantify the two main components of productivity, namely the fraction of active ribosomes and the number of synthesizing cycles. Here, we provide a direct and highly reliable measure of ribosomal activity in any given CFPS system, introducing an enhanced-arrest peptide variant. We observe an almost complete stalling of ribosomes that produce GFPem (~95%), as determined by common centrifugation techniques and fluorescence correlation spectroscopy (FCS). Moreover, we thoroughly study the effect of different ribosomal modifications independently on activity and number of synthesizing cycles. Finally, employing two-colour coincidence detection and two-colour colocalisation microscopy, we demonstrate real-time access to key productivity parameters with minimal sample consumption on a single ribosome level.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

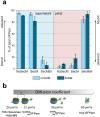
 and
and  without centrifugation (after 2 h CFPS reaction). Corresponding values for free GFPem and labelled 70S ribosomes are given for comparison. The intermediate value for
without centrifugation (after 2 h CFPS reaction). Corresponding values for free GFPem and labelled 70S ribosomes are given for comparison. The intermediate value for  reveals partial release, while the ribosome-like diffusion of
reveals partial release, while the ribosome-like diffusion of  indicates efficient stalling.
indicates efficient stalling.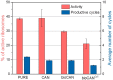
 (efficient stalling, only one productive cycle) and
(efficient stalling, only one productive cycle) and  (almost no stalling, constant activity assumed), respectively (after 2 h CFPS reaction). Activity drops with each modification step, while the number of synthesizing cycles remains mainly constant at a low level. The experiments were carried out at least 3 times for each condition (biological replications) in triplicates (technical replications).
(almost no stalling, constant activity assumed), respectively (after 2 h CFPS reaction). Activity drops with each modification step, while the number of synthesizing cycles remains mainly constant at a low level. The experiments were carried out at least 3 times for each condition (biological replications) in triplicates (technical replications).
 (right) images were acquired simultaneously at the same sample area. Colocalised ribosome (8 out of 49) and GFPem (8 out of 10) signals are marked (yellow circles). Scalebar: 5 μm.
(right) images were acquired simultaneously at the same sample area. Colocalised ribosome (8 out of 49) and GFPem (8 out of 10) signals are marked (yellow circles). Scalebar: 5 μm.Similar articles
-
Profiling of gene-dependent translational progress in cell-free protein synthesis by real-space imaging.Anal Biochem. 2009 Nov 15;394(2):275-80. doi: 10.1016/j.ab.2009.07.033. Epub 2009 Jul 28. Anal Biochem. 2009. PMID: 19643072
-
Cell-free compartmentalized protein synthesis inside double emulsion templated liposomes with in vitro synthesized and assembled ribosomes.Chem Commun (Camb). 2016 Apr 7;52(31):5467-9. doi: 10.1039/c6cc00223d. Chem Commun (Camb). 2016. PMID: 27019994 Free PMC article.
-
Single-molecule imaging of full protein synthesis by immobilized ribosomes.Nucleic Acids Res. 2008 Jul;36(12):e70. doi: 10.1093/nar/gkn338. Epub 2008 May 29. Nucleic Acids Res. 2008. PMID: 18511463 Free PMC article.
-
Inhibition by suramin of protein synthesis in vitro. Ribosomes as the target of the drug.Biochimie. 2006 May;88(5):497-503. doi: 10.1016/j.biochi.2005.10.009. Epub 2005 Nov 10. Biochimie. 2006. PMID: 16386828
-
[A model for trans-translation].Yi Chuan. 2006 Aug;28(8):1051-4. Yi Chuan. 2006. PMID: 16870596 Review. Chinese.
Cited by
-
Impact of Molecule Concentration, Diffusion Rates and Surface Passivation on Single-Molecule Fluorescence Studies in Solution.Biomolecules. 2022 Mar 18;12(3):468. doi: 10.3390/biom12030468. Biomolecules. 2022. PMID: 35327660 Free PMC article.
-
Distinct pre-initiation steps in human mitochondrial translation.Nat Commun. 2020 Jun 10;11(1):2932. doi: 10.1038/s41467-020-16503-2. Nat Commun. 2020. PMID: 32522994 Free PMC article.
-
Protein folding in vitro and in the cell: From a solitary journey to a team effort.Biophys Chem. 2022 Aug;287:106821. doi: 10.1016/j.bpc.2022.106821. Epub 2022 Apr 29. Biophys Chem. 2022. PMID: 35667131 Free PMC article. Review.
-
Brightness-gated two-color coincidence detection unravels two distinct mechanisms in bacterial protein translation initiation.Commun Biol. 2019 Dec 6;2:459. doi: 10.1038/s42003-019-0709-7. eCollection 2019. Commun Biol. 2019. PMID: 31840104 Free PMC article.
-
Cell-Free Protein Synthesis From Fast-Growing Vibrio natriegens.Front Microbiol. 2018 Jun 1;9:1146. doi: 10.3389/fmicb.2018.01146. eCollection 2018. Front Microbiol. 2018. PMID: 29910785 Free PMC article.
References
-
- Woolhead C. A., McCormick P. J. & Johnson A. E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

