Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities
- PMID: 28435450
- PMCID: PMC5399578
- DOI: 10.7150/thno.18551
Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities
Abstract
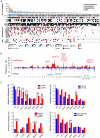
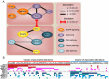
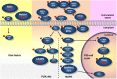
Oral squamous cell carcinoma (OSCC), an epithelial malignancy affecting a variety of subsites in the oral cavity, is prevalent in Asia. The survival rate of OSCC patients has not improved over the past decades due to its heterogeneous etiology, genetic aberrations, and treatment outcomes. Improvement in therapeutic strategies and tailored treatment options is an unmet need. To unveil the mutational spectrum, whole-exome sequencing of 120 OSCC from male individuals in Taiwan was conducted. Analyzing the contributions of the five mutational signatures extracted from the dataset of somatic variations identified four groups of tumors that were significantly associated with demographic and clinical features. In addition, known (TP53, FAT1, EPHA2, CDKN2A, NOTCH1, CASP8, HRAS, RASA1, and PIK3CA) and novel (CHUK and ELAVL1) genes that were significantly and frequently mutated in OSCC were discovered. Further analyses of gene alteration status with clinical parameters revealed that the tumors of the tongue were enriched with copy-number alterations in several gene clusters containing CCND1 and MAP4K2. Through defining the catalog of targetable genomic alterations, 58% of the tumors were found to carry at least one aberrant event potentially targeted by US Food and Drug Administration (FDA)-approved agents. Strikingly, if targeting the p53-cell cycle pathway (TP53 and CCND1) by the drugs studied in phase I-III clinical trials, those possibly actionable tumors are predominantly located in the tongue, suggesting a better prediction of sensitivity to current targeted therapies. Our work revealed molecular OSCC subgroups that reflect etiological and prognostic correlation as well as defined the landscape of major altered events in the coding regions of OSCC genomes. These findings provide clues for the design of clinical trials for targeted therapies and stratification of OSCC patients with differential therapeutic efficacy.
Keywords: Oral squamous cell carcinoma; driver gene; exome sequencing; mutational signature; targeted therapy..
Conflict of interest statement
Competing interest: The authors declare that they have no competing interests.
Figures

Similar articles
-
Exome sequencing of oral squamous cell carcinoma in users of Arabian snuff reveals novel candidates for driver genes.Int J Cancer. 2016 Jul 15;139(2):363-72. doi: 10.1002/ijc.30068. Epub 2016 Mar 18. Int J Cancer. 2016. PMID: 26934577 Free PMC article.
-
Precise Identification of Recurrent Somatic Mutations in Oral Cancer Through Whole-Exome Sequencing Using Multiple Mutation Calling Pipelines.Front Oncol. 2021 Nov 29;11:741626. doi: 10.3389/fonc.2021.741626. eCollection 2021. Front Oncol. 2021. PMID: 34912705 Free PMC article.
-
Genetically-defined novel oral squamous cell carcinoma cell lines for the development of molecular therapies.Oncotarget. 2016 May 10;7(19):27802-18. doi: 10.18632/oncotarget.8533. Oncotarget. 2016. PMID: 27050151 Free PMC article.
-
Translational genomics and recent advances in oral squamous cell carcinoma.Semin Cancer Biol. 2020 Apr;61:71-83. doi: 10.1016/j.semcancer.2019.09.011. Epub 2019 Sep 19. Semin Cancer Biol. 2020. PMID: 31542510 Review.
-
Targeting the DNA Damage Response in OSCC with TP53 Mutations.J Dent Res. 2018 Jun;97(6):635-644. doi: 10.1177/0022034518759068. Epub 2018 Feb 28. J Dent Res. 2018. PMID: 29489434 Free PMC article. Review.
Cited by
-
Systemic and Local Effects Among Patients With Betel Quid-Related Oral Cancer.Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221146870. doi: 10.1177/15330338221146870. Technol Cancer Res Treat. 2022. PMID: 36575633 Free PMC article. Review.
-
High Level of Plasma EGFL6 Is Associated with Clinicopathological Characteristics in Patients with Oral Squamous Cell Carcinoma.Int J Med Sci. 2017 Apr 8;14(5):419-424. doi: 10.7150/ijms.18555. eCollection 2017. Int J Med Sci. 2017. PMID: 28539817 Free PMC article.
-
Genomic landscape and clonal architecture of mouse oral squamous cell carcinomas dictate tumour ecology.Nat Commun. 2020 Nov 9;11(1):5671. doi: 10.1038/s41467-020-19401-9. Nat Commun. 2020. PMID: 33168804 Free PMC article.
-
Cotargeting CHK1 and PI3K Synergistically Suppresses Tumor Growth of Oral Cavity Squamous Cell Carcinoma in Patient-Derived Xenografts.Cancers (Basel). 2020 Jun 29;12(7):1726. doi: 10.3390/cancers12071726. Cancers (Basel). 2020. PMID: 32610557 Free PMC article.
-
The prognostic role of combining Krüppel-like factor 4 score and grade of inflammation in a Norwegian cohort of oral tongue squamous cell carcinomas.Eur J Oral Sci. 2022 Jun;130(3):e12866. doi: 10.1111/eos.12866. Epub 2022 Apr 1. Eur J Oral Sci. 2022. PMID: 35363406 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T. et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Zini A, Czerninski R, Sgan-Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med. 2010;39:299–305. - PubMed
-
- Krishna Rao SV, Mejia G, Roberts-Thomson K, Logan R. Epidemiology of oral cancer in Asia in the past decade-an update (2000-2012) Asian Pac J Cancer Prev. 2013;14:5567–77. - PubMed
-
- Su CC, Yang HF, Huang SJ, Lian Ie B. Distinctive features of oral cancer in Changhua County: high incidence, buccal mucosa preponderance, and a close relation to betel quid chewing habit. J Formos Med Assoc. 2007;106:225–33. - PubMed
-
- Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45:301–8. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

