Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation
- PMID: 28388414
- PMCID: PMC5388891
- DOI: 10.1016/j.cell.2017.03.031
Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation
Abstract
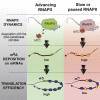
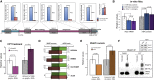
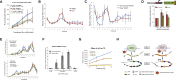
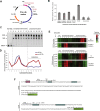
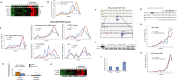
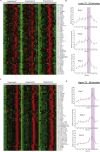
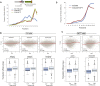
Transcription and translation are two main pillars of gene expression. Due to the different timings, spots of action, and mechanisms of regulation, these processes are mainly regarded as distinct and generally uncoupled, despite serving a common purpose. Here, we sought for a possible connection between transcription and translation. Employing an unbiased screen of multiple human promoters, we identified a positive effect of TATA box on translation and a general coupling between mRNA expression and translational efficiency. Using a CRISPR-Cas9-mediated approach, genome-wide analyses, and in vitro experiments, we show that the rate of transcription regulates the efficiency of translation. Furthermore, we demonstrate that m6A modification of mRNAs is co-transcriptional and depends upon the dynamics of the transcribing RNAPII. Suboptimal transcription rates lead to elevated m6A content, which may result in reduced translation. This study uncovers a general and widespread link between transcription and translation that is governed by epigenetic modification of mRNAs.
Keywords: N6-adenosine methylation; RNAPII; TATA; gene regulation; m(6)A; transcription; translation efficiency.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Dynamic m(6)A mRNA methylation directs translational control of heat shock response.Nature. 2015 Oct 22;526(7574):591-4. doi: 10.1038/nature15377. Epub 2015 Oct 12. Nature. 2015. PMID: 26458103 Free PMC article.
-
N6-methyladenosine is required for the hypoxic stabilization of specific mRNAs.RNA. 2017 Sep;23(9):1444-1455. doi: 10.1261/rna.061044.117. Epub 2017 Jun 13. RNA. 2017. PMID: 28611253 Free PMC article.
-
RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation.Genes Dev. 2015 Jul 1;29(13):1343-55. doi: 10.1101/gad.262766.115. Genes Dev. 2015. PMID: 26159994 Free PMC article. Review.
-
Post-transcriptional gene regulation by mRNA modifications.Nat Rev Mol Cell Biol. 2017 Jan;18(1):31-42. doi: 10.1038/nrm.2016.132. Epub 2016 Nov 3. Nat Rev Mol Cell Biol. 2017. PMID: 27808276 Free PMC article. Review.
-
Regulatory roles of N6-methyladenosine (m6A) methylation in RNA processing and non-communicable diseases.Cancer Gene Ther. 2024 Oct;31(10):1439-1453. doi: 10.1038/s41417-024-00789-1. Epub 2024 Jun 5. Cancer Gene Ther. 2024. PMID: 38839892 Review.
Cited by
-
Refined RIP-seq protocol for epitranscriptome analysis with low input materials.PLoS Biol. 2018 Sep 13;16(9):e2006092. doi: 10.1371/journal.pbio.2006092. eCollection 2018 Sep. PLoS Biol. 2018. PMID: 30212448 Free PMC article.
-
It's complicated… m6A-dependent regulation of gene expression in cancer.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):382-393. doi: 10.1016/j.bbagrm.2018.09.010. Epub 2018 Oct 5. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30296493 Free PMC article. Review.
-
circINSR Inhibits Adipogenic Differentiation of Adipose-Derived Stromal Vascular Fractions through the miR-152/MEOX2 Axis in Sheep.Int J Mol Sci. 2023 Feb 9;24(4):3501. doi: 10.3390/ijms24043501. Int J Mol Sci. 2023. PMID: 36834919 Free PMC article.
-
The N6-Methyladenosine Modification and Its Role in mRNA Metabolism and Gastrointestinal Tract Disease.Front Surg. 2022 Jan 28;9:819335. doi: 10.3389/fsurg.2022.819335. eCollection 2022. Front Surg. 2022. PMID: 35155557 Free PMC article. Review.
-
Epigenetic perspectives of COVID-19: Virus infection to disease progression and therapeutic control.Biochim Biophys Acta Mol Basis Dis. 2022 Dec 1;1868(12):166527. doi: 10.1016/j.bbadis.2022.166527. Epub 2022 Aug 22. Biochim Biophys Acta Mol Basis Dis. 2022. PMID: 36002132 Free PMC article.
References
-
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

