The Mutational Landscape of Circulating Tumor Cells in Multiple Myeloma
- PMID: 28380360
- PMCID: PMC5439509
- DOI: 10.1016/j.celrep.2017.03.025
The Mutational Landscape of Circulating Tumor Cells in Multiple Myeloma
Abstract
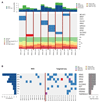
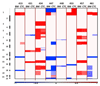
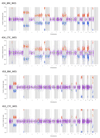
The development of sensitive and non-invasive "liquid biopsies" presents new opportunities for longitudinal monitoring of tumor dissemination and clonal evolution. The number of circulating tumor cells (CTCs) is prognostic in multiple myeloma (MM), but there is little information on their genetic features. Here, we have analyzed the genomic landscape of CTCs from 29 MM patients, including eight cases with matched/paired bone marrow (BM) tumor cells. Our results show that 100% of clonal mutations in patient BM were detected in CTCs and that 99% of clonal mutations in CTCs were present in BM MM. These include typical driver mutations in MM such as in KRAS, NRAS, or BRAF. These data suggest that BM and CTC samples have similar clonal structures, as discordances between the two were restricted to subclonal mutations. Accordingly, our results pave the way for potentially less invasive mutation screening of MM patients through characterization of CTCs.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates.Nat Commun. 2017 May 11;8:15086. doi: 10.1038/ncomms15086. Nat Commun. 2017. PMID: 28492226 Free PMC article. Clinical Trial.
-
Construction of a reference material panel for detecting KRAS/NRAS/EGFR/BRAF/MET mutations in plasma ctDNA.J Clin Pathol. 2021 May;74(5):314-320. doi: 10.1136/jclinpath-2020-206745. Epub 2020 Aug 17. J Clin Pathol. 2021. PMID: 32817175 Free PMC article.
-
Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer.Oncotarget. 2016 Dec 20;7(51):85349-85364. doi: 10.18632/oncotarget.13350. Oncotarget. 2016. PMID: 27863403 Free PMC article.
-
Tracking myeloma tumor DNA in peripheral blood.Best Pract Res Clin Haematol. 2020 Mar;33(1):101146. doi: 10.1016/j.beha.2020.101146. Epub 2020 Jan 14. Best Pract Res Clin Haematol. 2020. PMID: 32139012 Free PMC article. Review.
-
[Chromosomal abnormalities and clonal evolution in multiple myeloma].Rinsho Ketsueki. 2014 Oct;55(10):2036-45. Rinsho Ketsueki. 2014. PMID: 25297769 Review. Japanese. No abstract available.
Cited by
-
Cell-free DNA for genomic profiling and minimal residual disease monitoring in Myeloma- are we there yet?Am J Blood Res. 2020 Jun 15;10(3):26-45. eCollection 2020. Am J Blood Res. 2020. PMID: 32685257 Free PMC article. Review.
-
Transcriptional profiling of circulating tumor cells in multiple myeloma: a new model to understand disease dissemination.Leukemia. 2020 Feb;34(2):589-603. doi: 10.1038/s41375-019-0588-4. Epub 2019 Oct 8. Leukemia. 2020. PMID: 31595039
-
Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma.Cancers (Basel). 2022 Aug 26;14(17):4136. doi: 10.3390/cancers14174136. Cancers (Basel). 2022. PMID: 36077672 Free PMC article. Review.
-
Clinical application and detection techniques of liquid biopsy in gastric cancer.Mol Cancer. 2023 Jan 11;22(1):7. doi: 10.1186/s12943-023-01715-z. Mol Cancer. 2023. PMID: 36627698 Free PMC article. Review.
-
Ultradeep, targeted sequencing reveals distinct mutations in blood compared to matched bone marrow among patients with multiple myeloma.Blood Cancer J. 2019 Sep 30;9(10):77. doi: 10.1038/s41408-019-0238-0. Blood Cancer J. 2019. PMID: 31570697 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

