La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs
- PMID: 28379136
- PMCID: PMC5419741
- DOI: 10.7554/eLife.24146
La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs
Abstract
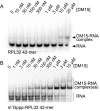
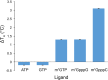
The 5'terminal oligopyrimidine (5'TOP) motif is a cis-regulatory RNA element located immediately downstream of the 7-methylguanosine [m7G] cap of TOP mRNAs, which encode ribosomal proteins and translation factors. In eukaryotes, this motif coordinates the synchronous and stoichiometric expression of the protein components of the translation machinery. La-related protein 1 (LARP1) binds TOP mRNAs, regulating their stability and translation. We present crystal structures of the human LARP1 DM15 region in complex with a 5'TOP motif, a cap analog (m7GTP), and a capped cytidine (m7GpppC), resolved to 2.6, 1.8 and 1.7 Å, respectively. Our binding, competition, and immunoprecipitation data corroborate and elaborate on the mechanism of 5'TOP motif binding by LARP1. We show that LARP1 directly binds the cap and adjacent 5'TOP motif of TOP mRNAs, effectively impeding access of eIF4E to the cap and preventing eIF4F assembly. Thus, LARP1 is a specialized TOP mRNA cap-binding protein that controls ribosome biogenesis.
Keywords: 5' cap; E. coli; La-related protein 1; RNA binding protein; TOP mRNAs; X-ray crystallography; biochemistry; biophysics; eIF4E; human; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



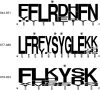





Similar articles
-
La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1).J Biol Chem. 2015 Jun 26;290(26):15996-6020. doi: 10.1074/jbc.M114.621730. Epub 2015 May 4. J Biol Chem. 2015. PMID: 25940091 Free PMC article.
-
The LARP1 homolog Slr1p controls the stability and expression of proto-5'TOP mRNAs in fission yeast.Cell Rep. 2023 Oct 31;42(10):113226. doi: 10.1016/j.celrep.2023.113226. Epub 2023 Oct 17. Cell Rep. 2023. PMID: 37851576
-
The amino acid sensor GCN2 suppresses terminal oligopyrimidine (TOP) mRNA translation via La-related protein 1 (LARP1).J Biol Chem. 2022 Sep;298(9):102277. doi: 10.1016/j.jbc.2022.102277. Epub 2022 Jul 19. J Biol Chem. 2022. PMID: 35863436 Free PMC article.
-
LARP1 and LARP4: up close with PABP for mRNA 3' poly(A) protection and stabilization.RNA Biol. 2021 Feb;18(2):259-274. doi: 10.1080/15476286.2020.1868753. Epub 2021 Jan 31. RNA Biol. 2021. PMID: 33522422 Free PMC article. Review.
-
LARP1 on TOP of ribosome production.Wiley Interdiscip Rev RNA. 2018 Sep;9(5):e1480. doi: 10.1002/wrna.1480. Epub 2018 May 2. Wiley Interdiscip Rev RNA. 2018. PMID: 29722158 Free PMC article. Review.
Cited by
-
The Translational Regulation in mTOR Pathway.Biomolecules. 2022 Jun 8;12(6):802. doi: 10.3390/biom12060802. Biomolecules. 2022. PMID: 35740927 Free PMC article. Review.
-
La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region.Nucleic Acids Res. 2018 Feb 16;46(3):1457-1469. doi: 10.1093/nar/gkx1237. Nucleic Acids Res. 2018. PMID: 29244122 Free PMC article.
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells.Genome Biol. 2024 Jan 23;25(1):31. doi: 10.1186/s13059-023-03146-x. Genome Biol. 2024. PMID: 38263082 Free PMC article. Review.
-
RNA localization mechanisms transcend cell morphology.Elife. 2023 Mar 3;12:e80040. doi: 10.7554/eLife.80040. Elife. 2023. PMID: 36867563 Free PMC article.
-
Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease.Life Sci. 2018 Nov 1;212:138-144. doi: 10.1016/j.lfs.2018.09.054. Epub 2018 Oct 2. Life Sci. 2018. PMID: 30290184 Free PMC article. Review.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5'-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Molecular and Cellular Biology. 1994;14:3822–3833. doi: 10.1128/MCB.14.6.3822. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

