CXCR1 remodels the vascular niche to promote hematopoietic stem and progenitor cell engraftment
- PMID: 28351983
- PMCID: PMC5379982
- DOI: 10.1084/jem.20161616
CXCR1 remodels the vascular niche to promote hematopoietic stem and progenitor cell engraftment
Abstract
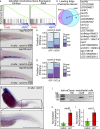
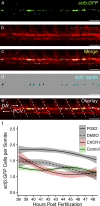
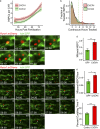
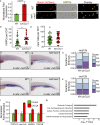
The microenvironment is an important regulator of hematopoietic stem and progenitor cell (HSPC) biology. Recent advances marking fluorescent HSPCs have allowed exquisite visualization of HSPCs in the caudal hematopoietic tissue (CHT) of the developing zebrafish. Here, we show that the chemokine cxcl8 and its receptor, cxcr1, are expressed by zebrafish endothelial cells, and we identify cxcl8/cxcr1 signaling as a positive regulator of HSPC colonization. Single-cell tracking experiments demonstrated that this is a result of increases in HSPC-endothelial cell "cuddling," HSPC residency time within the CHT, and HSPC mitotic rate. Enhanced cxcl8/cxcr1 signaling was associated with an increase in the volume of the CHT and induction of cxcl12a expression. Finally, using parabiotic zebrafish, we show that cxcr1 acts HSPC nonautonomously to improve the efficiency of donor HSPC engraftment. This work identifies a mechanism by which the hematopoietic niche remodels to promote HSPC engraftment and suggests that cxcl8/cxcr1 signaling is a potential therapeutic target in patients undergoing hematopoietic stem cell transplantation.
© 2017 Blaser et al.
Figures



Similar articles
-
The Vascular Niche Regulates Hematopoietic Stem and Progenitor Cell Lodgment and Expansion via klf6a-ccl25b.Dev Cell. 2017 Aug 21;42(4):349-362.e4. doi: 10.1016/j.devcel.2017.07.012. Epub 2017 Aug 10. Dev Cell. 2017. PMID: 28803829
-
A single-cell resolution developmental atlas of hematopoietic stem and progenitor cell expansion in zebrafish.Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2015748118. doi: 10.1073/pnas.2015748118. Proc Natl Acad Sci U S A. 2021. PMID: 33785593 Free PMC article.
-
Non-fucosylated CB CD34+ cells represent a good target for enforced fucosylation to improve engraftment following cord blood transplantation.Cytotherapy. 2017 Feb;19(2):285-292. doi: 10.1016/j.jcyt.2016.11.001. Epub 2016 Dec 2. Cytotherapy. 2017. PMID: 27919572
-
Advances in umbilical cord blood stem cell expansion and clinical translation.Exp Hematol. 2015 Jul;43(7):498-513. doi: 10.1016/j.exphem.2015.04.011. Epub 2015 May 10. Exp Hematol. 2015. PMID: 25970610 Review.
-
Hematopoietic Niche - Exploring Biomimetic Cues to Improve the Functionality of Hematopoietic Stem/Progenitor Cells.Biotechnol J. 2018 Feb;13(2). doi: 10.1002/biot.201700088. Epub 2017 Dec 28. Biotechnol J. 2018. PMID: 29178199 Review.
Cited by
-
Molecular and Cellular Mechanisms of Vascular Development in Zebrafish.Life (Basel). 2021 Oct 15;11(10):1088. doi: 10.3390/life11101088. Life (Basel). 2021. PMID: 34685459 Free PMC article. Review.
-
Niches that regulate stem cells and hematopoiesis in adult bone marrow.Dev Cell. 2021 Jul 12;56(13):1848-1860. doi: 10.1016/j.devcel.2021.05.018. Epub 2021 Jun 18. Dev Cell. 2021. PMID: 34146467 Free PMC article. Review.
-
Transcription factor induction of vascular blood stem cell niches in vivo.Dev Cell. 2023 Jun 19;58(12):1037-1051.e4. doi: 10.1016/j.devcel.2023.04.007. Epub 2023 Apr 28. Dev Cell. 2023. PMID: 37119815 Free PMC article.
-
[The pathogenesis of poor graft function after allogeneic hematopoietic stem cell transplantation].Zhonghua Xue Ye Xue Za Zhi. 2019 Sep 14;40(9):792-795. doi: 10.3760/cma.j.issn.0253-2727.2019.09.021. Zhonghua Xue Ye Xue Za Zhi. 2019. PMID: 31648490 Free PMC article. Chinese. No abstract available.
-
Location, Location, Location: How Vascular Specialization Influences Hematopoietic Fates During Development.Front Cell Dev Biol. 2020 Nov 13;8:602617. doi: 10.3389/fcell.2020.602617. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33282876 Free PMC article. Review.
References
-
- Butler J.M., Nolan D.J., Vertes E.L., Varnum-Finney B., Kobayashi H., Hooper A.T., Seandel M., Shido K., White I.A., Kobayashi M., et al. . 2010. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 6:251–264. 10.1016/j.stem.2010.02.001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

