INSPIIRED: A Pipeline for Quantitative Analysis of Sites of New DNA Integration in Cellular Genomes
- PMID: 28344990
- PMCID: PMC5363316
- DOI: 10.1016/j.omtm.2016.11.002
INSPIIRED: A Pipeline for Quantitative Analysis of Sites of New DNA Integration in Cellular Genomes
Abstract
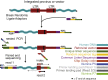
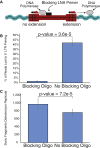
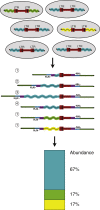
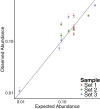
Integration of new DNA into cellular genomes mediates replication of retroviruses and transposons; integration reactions have also been adapted for use in human gene therapy. Tracking the distributions of integration sites is important to characterize populations of transduced cells and to monitor potential outgrow of pathogenic cell clones. Here, we describe a pipeline for quantitative analysis of integration site distributions named INSPIIRED (integration site pipeline for paired-end reads). We describe optimized biochemical steps for site isolation using Illumina paired-end sequencing, including new technology for suppressing recovery of unwanted contaminants, then software for alignment, quality control, and management of integration site sequences. During library preparation, DNAs are broken by sonication, so that after ligation-mediated PCR the number of ligation junction sites can be used to infer abundance of gene-modified cells. We generated integration sites of known positions in silico, and we describe optimization of sample processing parameters refined by comparison to truth. We also present a novel graph-theory-based method for quantifying integration sites in repeated sequences, and we characterize the consequences using synthetic and experimental data. In an accompanying paper, we describe an additional set of statistical tools for data analysis and visualization. Software is available at https://github.com/BushmanLab/INSPIIRED.
Keywords: SCID-X1; gammaretrovirus; gene therapy; insertional mutagenesis; lentivirus; mutagenesis; recombination; retrovirus; vector; vector driving.
Figures

Similar articles
-
INSPIIRED: Quantification and Visualization Tools for Analyzing Integration Site Distributions.Mol Ther Methods Clin Dev. 2016 Dec 18;4:17-26. doi: 10.1016/j.omtm.2016.11.003. eCollection 2017 Mar 17. Mol Ther Methods Clin Dev. 2016. PMID: 28344988 Free PMC article.
-
LUMI-PCR: an Illumina platform ligation-mediated PCR protocol for integration site cloning, provides molecular quantitation of integration sites.Mob DNA. 2020 Feb 4;11:7. doi: 10.1186/s13100-020-0201-4. eCollection 2020. Mob DNA. 2020. PMID: 32042315 Free PMC article.
-
VISPA2: a scalable pipeline for high-throughput identification and annotation of vector integration sites.BMC Bioinformatics. 2017 Nov 25;18(1):520. doi: 10.1186/s12859-017-1937-9. BMC Bioinformatics. 2017. PMID: 29178837 Free PMC article.
-
Retroviral Insertional Mutagenesis in Humans: Evidence for Four Genetic Mechanisms Promoting Expansion of Cell Clones.Mol Ther. 2020 Feb 5;28(2):352-356. doi: 10.1016/j.ymthe.2019.12.009. Epub 2020 Jan 7. Mol Ther. 2020. PMID: 31951833 Free PMC article. Review.
-
Integrating Vectors for Gene Therapy and Clonal Tracking of Engineered Hematopoiesis.Hematol Oncol Clin North Am. 2017 Oct;31(5):737-752. doi: 10.1016/j.hoc.2017.06.009. Hematol Oncol Clin North Am. 2017. PMID: 28895844 Review.
Cited by
-
T cell dynamics and response of the microbiota after gene therapy to treat X-linked severe combined immunodeficiency.Genome Med. 2018 Sep 28;10(1):70. doi: 10.1186/s13073-018-0580-z. Genome Med. 2018. PMID: 30261899 Free PMC article.
-
HIV integration in the human brain is linked to microglial activation and 3D genome remodeling.Mol Cell. 2022 Dec 15;82(24):4647-4663.e8. doi: 10.1016/j.molcel.2022.11.016. Mol Cell. 2022. PMID: 36525955 Free PMC article.
-
Development of an in vitro genotoxicity assay to detect retroviral vector-induced lymphoid insertional mutants.Mol Ther Methods Clin Dev. 2023 Aug 22;30:515-533. doi: 10.1016/j.omtm.2023.08.017. eCollection 2023 Sep 14. Mol Ther Methods Clin Dev. 2023. PMID: 37693949 Free PMC article.
-
Meeting FDA Guidance recommendations for replication-competent virus and insertional oncogenesis testing.Mol Ther Methods Clin Dev. 2022 Dec 2;28:28-39. doi: 10.1016/j.omtm.2022.11.009. eCollection 2023 Mar 9. Mol Ther Methods Clin Dev. 2022. PMID: 36588821 Free PMC article. Review.
-
SeqVerify: An accessible analysis tool for cell line genomic integrity, contamination, and gene editing outcomes.Stem Cell Reports. 2024 Oct 8;19(10):1505-1515. doi: 10.1016/j.stemcr.2024.08.004. Epub 2024 Sep 12. Stem Cell Reports. 2024. PMID: 39270651 Free PMC article.
References
-
- Bushman F.D. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Lateral DNA Transfer: Mechanisms and Consequences.
-
- Craig N.L., Craigie R., Gellert M., Lambowitz A.M. American Society for Microbiology Press; Washington, D.C.: 2002. Mobile DNA II.
-
- Schröder A.R.W., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. - PubMed
-
- Coffin J.M., Hughes S.H., Varmus H.E. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. Retroviruses. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

