Effects of miR-145-5p through NRAS on the cell proliferation, apoptosis, migration, and invasion in melanoma by inhibiting MAPK and PI3K/AKT pathways
- PMID: 28332309
- PMCID: PMC5387172
- DOI: 10.1002/cam4.1030
Effects of miR-145-5p through NRAS on the cell proliferation, apoptosis, migration, and invasion in melanoma by inhibiting MAPK and PI3K/AKT pathways
Retraction in
-
Retraction.Cancer Med. 2023 Feb;12(4):5173. doi: 10.1002/cam4.5232. Epub 2022 Sep 8. Cancer Med. 2023. PMID: 36081336 Free PMC article. No abstract available.
Abstract
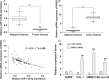
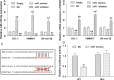
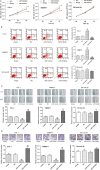
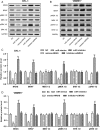
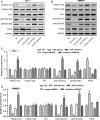
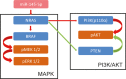
We aimed to detect the effects of miR-145-5p on the cell proliferation, apoptosis, migration, and invasion in NRAS-mutant, BRAF-mutant, and wild-type melanoma cells, in order to figure out the potential mechanisms and provide a novel therapeutic target of melanoma. RT-qPCR and western blot were used to detect the expression of miR-145-5p and NRAS in melanoma tumor tissues and cells, respectively. Luciferase assay was performed to determine whether miR-145-5p directly targeted NRAS. After transfecting miR-145-5p mimics, miR-145-5p inhibitors, NRAS cDNA and NRAS siRNA into CHL-1, VMM917 and SK-mel-28 cells, functional assays were used to detect the proliferation, apoptosis, invasion and migration, including MTT, flow cytometry, Transwell and wound healing assays. In addition, xenograft models in nude mice were also conducted to verify the role of miR-145-5p in vivo. MiR-145-5p was able to suppress proliferation, invasion, and migration of VMM917 and CHL-1 cells and induce apoptosis by inhibiting MAPK and PI3K/AKT pathways. However, aberrant expression of miR-145-5p and NRAS has little impact on the viability and metastasis of BRAF-mutant melanoma. The higher expression of miR-145-5p in xenograft models repressed the VMM917-induced and CHL-1-induced tumor growth observably and has little effect on SK-mel-28-induced tumor growth which was consistent with the results in vitro. Through targeting NRAS, miR-145-5p could suppress cell proliferation, invasion, and migration and induce apoptosis of CHL-1 and VMM917 melanoma cells by inhibiting MAPK and PI3K/AKT pathways.
Keywords: MAPK; NRAS; melanoma; miR-145-5p.
© 2017 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures


Similar articles
-
miR-224-5p inhibits proliferation, migration, and invasion by targeting PIK3R3/AKT3 in uveal melanoma.J Cell Biochem. 2019 Aug;120(8):12412-12421. doi: 10.1002/jcb.28507. Epub 2019 Mar 1. J Cell Biochem. 2019. PMID: 30825222
-
MicroRNA-98-5p modulates cervical cancer progression via controlling PI3K/AKT pathway.Bioengineered. 2021 Dec;12(2):10596-10607. doi: 10.1080/21655979.2021.2000722. Bioengineered. 2021. PMID: 34895048 Free PMC article.
-
MiR-592 Promotes Gastric Cancer Proliferation, Migration, and Invasion Through the PI3K/AKT and MAPK/ERK Signaling Pathways by Targeting Spry2.Cell Physiol Biochem. 2018;47(4):1465-1481. doi: 10.1159/000490839. Epub 2018 Jun 21. Cell Physiol Biochem. 2018. PMID: 29949784
-
Effects of MIR143 on rat sarcoma signaling networks in solid tumors: A brief overview.Cancer Sci. 2020 Apr;111(4):1076-1083. doi: 10.1111/cas.14357. Epub 2020 Mar 18. Cancer Sci. 2020. PMID: 32077199 Free PMC article. Review.
-
Molecular biology of microRNA-342 during tumor progression and invasion.Pathol Res Pract. 2023 Aug;248:154672. doi: 10.1016/j.prp.2023.154672. Epub 2023 Jul 3. Pathol Res Pract. 2023. PMID: 37413875 Review.
Cited by
-
MicroRNA-145-5p aggravates cell apoptosis and oxidative stress in tongue squamous cell carcinoma.Exp Ther Med. 2021 Apr;21(4):373. doi: 10.3892/etm.2021.9804. Epub 2021 Feb 19. Exp Ther Med. 2021. PMID: 33732346 Free PMC article.
-
Peptide Targeted Gold Nanoplatform Carrying miR-145 Induces Antitumoral Effects in Ovarian Cancer Cells.Pharmaceutics. 2022 Apr 28;14(5):958. doi: 10.3390/pharmaceutics14050958. Pharmaceutics. 2022. PMID: 35631544 Free PMC article.
-
Muscone suppresses gastric cancer via regulation of miRNA-145.Food Sci Nutr. 2021 Jul 26;9(9):4711-4721. doi: 10.1002/fsn3.2269. eCollection 2021 Sep. Food Sci Nutr. 2021. PMID: 34531985 Free PMC article.
-
Clinical significance of miR-1298 in cervical cancer and its biological function in vitro.Oncol Lett. 2021 May;21(5):401. doi: 10.3892/ol.2021.12662. Epub 2021 Mar 18. Oncol Lett. 2021. Retraction in: Oncol Lett. 2024 Feb 12;27(4):150. doi: 10.3892/ol.2024.14283. PMID: 33777224 Free PMC article. Retracted.
-
LncRNA CASC9 regulates cell proliferation, apoptosis and cell cycle via sponging miR-145-5p in colon cancer cells.Transl Cancer Res. 2019 Dec;8(8):2769-2780. doi: 10.21037/tcr.2019.10.33. Transl Cancer Res. 2019. PMID: 35117034 Free PMC article.
References
-
- Paluncic, J. , Kovacevic Z., Jansson P. J., et al. 2016. Roads to melanoma: Key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim. Biophys. Acta 1863:770–784. - PubMed
-
- Maio, M . 2012. Melanoma as a model tumour for immuno‐oncology. Ann. Oncol. 23 (Suppl 8):viii10–viii14. - PubMed
-
- Siegel, R. L. , Miller K. D., and Jemal A.. 2016. Cancer statistics, 2016. CA Cancer J. Clin. 66:7–30. - PubMed
-
- Abildgaard, C. , and Guldberg P.. 2015. Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol. Med. 21:164–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

