Kaposi's Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II To Create a Viral Transcriptional Factory
- PMID: 28331082
- PMCID: PMC5432858
- DOI: 10.1128/JVI.02491-16
Kaposi's Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II To Create a Viral Transcriptional Factory
Abstract
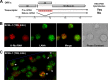
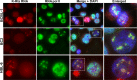
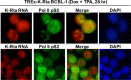
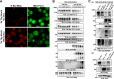
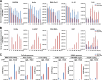
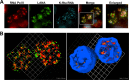
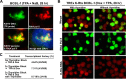
Locally concentrated nuclear factors ensure efficient binding to DNA templates, facilitating RNA polymerase II recruitment and frequent reutilization of stable preinitiation complexes. We have uncovered a mechanism for effective viral transcription by focal assembly of RNA polymerase II around Kaposi's sarcoma-associated herpesvirus (KSHV) genomes in the host cell nucleus. Using immunofluorescence labeling of latent nuclear antigen (LANA) protein, together with fluorescence in situ RNA hybridization (RNA-FISH) of the intron region of immediate early transcripts, we visualized active transcription of viral genomes in naturally infected cells. At the single-cell level, we found that not all episomes were uniformly transcribed following reactivation stimuli. However, those episomes that were being transcribed would spontaneously aggregate to form transcriptional "factories," which recruited a significant fraction of cellular RNA polymerase II. Focal assembly of "viral transcriptional factories" decreased the pool of cellular RNA polymerase II available for cellular gene transcription, which consequently impaired cellular gene expression globally, with the exception of selected ones. The viral transcriptional factories localized with replicating viral genomic DNAs. The observed colocalization of viral transcriptional factories with replicating viral genomic DNA suggests that KSHV assembles an "all-in-one" factory for both gene transcription and DNA replication. We propose that the assembly of RNA polymerase II around viral episomes in the nucleus may be a previously unexplored aspect of KSHV gene regulation by confiscation of a limited supply of RNA polymerase II in infected cells.IMPORTANCE B cells infected with Kaposi's sarcoma-associated herpesvirus (KSHV) harbor multiple copies of the KSHV genome in the form of episomes. Three-dimensional imaging of viral gene expression in the nucleus allows us to study interactions and changes in the physical distribution of these episomes following stimulation. The results showed heterogeneity in the responses of individual KSHV episomes to stimuli within a single reactivating cell; those episomes that did respond to stimulation, aggregated within large domains that appear to function as viral transcription factories. A significant portion of cellular RNA polymerase II was trapped in these factories and served to transcribe viral genomes, which coincided with an overall decrease in cellular gene expression. Our findings uncover a strategy of KSHV gene regulation through focal assembly of KSHV episomes and a molecular mechanism of late gene expression.
Keywords: KSHV; Kaposi's sarcoma-associated herpesvirus; RNA polymerase II; reactivation; regulation of gene expression; transcription; transcriptional factory.
Copyright © 2017 Chen et al.
Figures
Similar articles
-
Full-Length Isoforms of Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Accumulate in the Cytoplasm of Cells Undergoing the Lytic Cycle of Replication.J Virol. 2017 Nov 30;91(24):e01532-17. doi: 10.1128/JVI.01532-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978712 Free PMC article.
-
Sirtuin 6 Attenuates Kaposi's Sarcoma-Associated Herpesvirus Reactivation by Suppressing Ori-Lyt Activity and Expression of RTA.J Virol. 2019 Mar 21;93(7):e02200-18. doi: 10.1128/JVI.02200-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651359 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus terminal repeat regulates inducible lytic gene promoters.J Virol. 2024 Feb 20;98(2):e0138623. doi: 10.1128/jvi.01386-23. Epub 2024 Jan 19. J Virol. 2024. PMID: 38240593 Free PMC article.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
-
Long non-coding RNA and epigenetic gene regulation of KSHV.Viruses. 2014 Nov 4;6(11):4165-77. doi: 10.3390/v6114165. Viruses. 2014. PMID: 25375882 Free PMC article. Review.
Cited by
-
Quantitative Proteomics Analysis of Lytic KSHV Infection in Human Endothelial Cells Reveals Targets of Viral Immune Modulation.Cell Rep. 2020 Oct 13;33(2):108249. doi: 10.1016/j.celrep.2020.108249. Cell Rep. 2020. PMID: 33053346 Free PMC article.
-
Transcriptional and post-transcriptional regulation of viral gene expression in the gamma-herpesvirus Kaposi's sarcoma-associated herpesvirus.Curr Clin Microbiol Rep. 2018 Dec;5(4):219-228. doi: 10.1007/s40588-018-0102-1. Epub 2018 Aug 3. Curr Clin Microbiol Rep. 2018. PMID: 30854283 Free PMC article.
-
A Panel of Kaposi's Sarcoma-Associated Herpesvirus Mutants in the Polycistronic Kaposin Locus for Precise Analysis of Individual Protein Products.J Virol. 2022 Mar 9;96(5):e0156021. doi: 10.1128/JVI.01560-21. Epub 2021 Dec 22. J Virol. 2022. PMID: 34936820 Free PMC article.
-
KSHV RTA utilizes the host E3 ubiquitin ligase complex RNF20/40 to drive lytic reactivation.J Virol. 2023 Nov 30;97(11):e0138923. doi: 10.1128/jvi.01389-23. Epub 2023 Oct 27. J Virol. 2023. PMID: 37888983 Free PMC article.
-
Epigenetic reprogramming of host and viral genes by Human Cytomegalovirus infection in Kasumi-3 myeloid progenitor cells at early times post-infection.J Virol. 2021 May 10;95(11):e00183-21. doi: 10.1128/JVI.00183-21. Epub 2021 Mar 17. J Virol. 2021. PMID: 33731453 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

