Limited understanding of the functional diversity of N-linked glycans as a major gap of prion biology
- PMID: 28324664
- PMCID: PMC5399891
- DOI: 10.1080/19336896.2017.1301338
Limited understanding of the functional diversity of N-linked glycans as a major gap of prion biology
Abstract
Among a broad range of hypotheses on the molecular nature of transmissible spongiform encephalopathy or scrapie agents discussed in 1960s was a hypothesis of self-replicating polysaccharides. While the studies of the past 40 years provided unambiguous proof that this is not the case, emerging evidence suggests that carbohydrates in the form of sialylated N-linked glycans, which are a constitutive part of mammalian prions or PrPSc, are essential in determining prion fate in an organism. The current extra-view article discusses recent advancements on the role of N-linked glycans and specifically their sialylation status in controlling prion fate. In addition, this manuscript introduces a new concept on the important role of strain-specific functional carbohydrate epitopes on the PrPSc surface as main determinants of strain-specific biologic features. According to this concept, individual strain-specific folding patterns of PrPSc govern selection of PrPC sialoglycoforms expressed by a host that can be accommodated within particular PrPSc structures. Strain-specific patterns of functional carbohydrate epitopes formed by N-linked glycans on PrPSc surfaces define strain-specific biologic features. As a constitutive part of PrPSc, the individual strain-specific patterns of carbohydrate epitopes propagate faithfully within a given host as long as individual strain-specific PrPSc structures are maintained, ensuring inheritance of strain-specific biologic features.
Keywords: N-linked glycans; carbohydrate epitopes; microglia; prion; prion diseases; secondary lymphoid organs; sialic acid; sialylation.
Figures
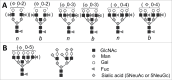
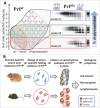
Comment on
- Extra View to: Srivastava S, Katorcha E, Daus ML, Lasch P, Beekes M, Baskakov IV. Sialylation controls prion fate in vivo. J Biol Chem 2017; 292(6): 2359-2368; http://dx.doi.org/10.1074/jbc.M116.768010
Similar articles
-
Region-Specific Sialylation Pattern of Prion Strains Provides Novel Insight into Prion Neurotropism.Int J Mol Sci. 2020 Jan 28;21(3):828. doi: 10.3390/ijms21030828. Int J Mol Sci. 2020. PMID: 32012886 Free PMC article.
-
Role of sialylation of N-linked glycans in prion pathogenesis.Cell Tissue Res. 2023 Apr;392(1):201-214. doi: 10.1007/s00441-022-03584-2. Epub 2022 Jan 28. Cell Tissue Res. 2023. PMID: 35088180 Free PMC article. Review.
-
Prion Strain-Specific Structure and Pathology: A View from the Perspective of Glycobiology.Viruses. 2018 Dec 18;10(12):723. doi: 10.3390/v10120723. Viruses. 2018. PMID: 30567302 Free PMC article. Review.
-
Role of sialylation in prion disease pathogenesis and prion structure.Prog Mol Biol Transl Sci. 2020;175:31-52. doi: 10.1016/bs.pmbts.2020.07.004. Epub 2020 Aug 24. Prog Mol Biol Transl Sci. 2020. PMID: 32958238 Review.
-
Analyses of N-linked glycans of PrPSc revealed predominantly 2,6-linked sialic acid residues.FEBS J. 2017 Nov;284(21):3727-3738. doi: 10.1111/febs.14268. Epub 2017 Sep 30. FEBS J. 2017. PMID: 28898525 Free PMC article.
Cited by
-
Region-specific glial homeostatic signature in prion diseases is replaced by a uniform neuroinflammation signature, common for brain regions and prion strains with different cell tropism.Neurobiol Dis. 2020 Apr;137:104783. doi: 10.1016/j.nbd.2020.104783. Epub 2020 Jan 27. Neurobiol Dis. 2020. PMID: 32001329 Free PMC article.
-
Inflammatory response of microglia to prions is controlled by sialylation of PrPSc.Sci Rep. 2018 Jul 27;8(1):11326. doi: 10.1038/s41598-018-29720-z. Sci Rep. 2018. PMID: 30054538 Free PMC article.
-
Region-Specific Sialylation Pattern of Prion Strains Provides Novel Insight into Prion Neurotropism.Int J Mol Sci. 2020 Jan 28;21(3):828. doi: 10.3390/ijms21030828. Int J Mol Sci. 2020. PMID: 32012886 Free PMC article.
-
Prion replication environment defines the fate of prion strain adaptation.PLoS Pathog. 2018 Jun 21;14(6):e1007093. doi: 10.1371/journal.ppat.1007093. eCollection 2018 Jun. PLoS Pathog. 2018. PMID: 29928047 Free PMC article.
-
Role of sialylation of N-linked glycans in prion pathogenesis.Cell Tissue Res. 2023 Apr;392(1):201-214. doi: 10.1007/s00441-022-03584-2. Epub 2022 Jan 28. Cell Tissue Res. 2023. PMID: 35088180 Free PMC article. Review.
References
-
- Field EJ. Transmission experiments with multiple sclerosis: an interim report. Br Med J 1966; 2:564-5; PMID:5950508; http://dx.doi.org/10.1136/bmj.2.5513.564 - DOI - PMC - PubMed
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136-44; PMID:6801762; http://dx.doi.org/10.1126/science.6801762 - DOI - PubMed
-
- Katorcha E, Makarava N, Savtchenko R, D'Azzo A, Baskakov IV. Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLOS Pathog 2014; 10:e1004366; PMID:25211026; http://dx.doi.org/10.1371/journal.ppat.1004366 - DOI - PMC - PubMed
-
- Srivastava S, Katorcha E, Daus ML, Lasch P, Beekes M, Baskakov IV. Sialylation controls prion fate in vivo. J Biol Chem 2017; 292:2359-68; PMID:27998976; http://dx.doi.org/10.1074/jbc.M116.768010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
