Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit
- PMID: 28264193
- PMCID: PMC5340527
- DOI: 10.7554/eLife.19594
Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit
Abstract
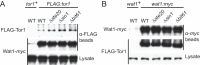
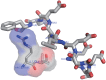
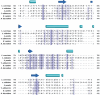
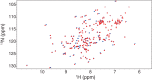
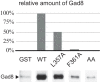
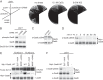
The target of rapamycin (TOR) protein kinase forms multi-subunit TOR complex 1 (TORC1) and TOR complex 2 (TORC2), which exhibit distinct substrate specificities. Sin1 is one of the TORC2-specific subunit essential for phosphorylation and activation of certain AGC-family kinases. Here, we show that Sin1 is dispensable for the catalytic activity of TORC2, but its conserved region in the middle (Sin1CRIM) forms a discrete domain that specifically binds the TORC2 substrate kinases. Sin1CRIM fused to a different TORC2 subunit can recruit the TORC2 substrate Gad8 for phosphorylation even in the sin1 null mutant of fission yeast. The solution structure of Sin1CRIM shows a ubiquitin-like fold with a characteristic acidic loop, which is essential for interaction with the TORC2 substrates. The specific substrate-recognition function is conserved in human Sin1CRIM, which may represent a potential target for novel anticancer drugs that prevent activation of the mTORC2 substrates such as AKT.
Keywords: AKT; S. pombe; SIN1; TOR complex 2; biochemistry; cell biology; human; substrate specificity; target of rapamycin.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures







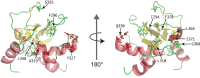






Comment in
-
A central role for a region in the middle.Elife. 2017 Mar 7;6:e25700. doi: 10.7554/eLife.25700. Elife. 2017. PMID: 28266914 Free PMC article.
Similar articles
-
(1)H, (15)N and (13)C resonance assignments of the conserved region in the middle domain of S. pombe Sin1 protein.Biomol NMR Assign. 2015 Apr;9(1):89-92. doi: 10.1007/s12104-014-9550-6. Epub 2014 Mar 8. Biomol NMR Assign. 2015. PMID: 24610629
-
TOR Complex 2- independent mutations in the regulatory PIF pocket of Gad8AKT1/SGK1 define separate branches of the stress response mechanisms in fission yeast.PLoS Genet. 2020 Nov 2;16(11):e1009196. doi: 10.1371/journal.pgen.1009196. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33137119 Free PMC article.
-
Modulation of TOR complex 2 signaling by the stress-activated MAPK pathway in fission yeast.J Cell Sci. 2019 Oct 10;132(19):jcs236133. doi: 10.1242/jcs.236133. J Cell Sci. 2019. PMID: 31477575
-
TORC1-Dependent Phosphorylation Targets in Fission Yeast.Biomolecules. 2017 Jul 3;7(3):50. doi: 10.3390/biom7030050. Biomolecules. 2017. PMID: 28671615 Free PMC article. Review.
-
Express yourself: how PP2A-B55Pab1 helps TORC1 talk to TORC2.Curr Genet. 2018 Feb;64(1):43-51. doi: 10.1007/s00294-017-0721-8. Epub 2017 Jun 22. Curr Genet. 2018. PMID: 28643116 Free PMC article. Review.
Cited by
-
New mechanistic insights into the RAS-SIN1 interaction at the membrane.Front Cell Dev Biol. 2022 Oct 6;10:987754. doi: 10.3389/fcell.2022.987754. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36274845 Free PMC article.
-
TOR complex 2 is a master regulator of plasma membrane homeostasis.Biochem J. 2022 Sep 30;479(18):1917-1940. doi: 10.1042/BCJ20220388. Biochem J. 2022. PMID: 36149412 Free PMC article.
-
The mTORC2 signaling network: targets and cross-talks.Biochem J. 2024 Jan 25;481(2):45-91. doi: 10.1042/BCJ20220325. Biochem J. 2024. PMID: 38270460 Free PMC article. Review.
-
mTOR-Rictor-EGFR axis in oncogenesis and diagnosis of glioblastoma multiforme.Mol Biol Rep. 2021 May;48(5):4813-4835. doi: 10.1007/s11033-021-06462-2. Epub 2021 Jun 16. Mol Biol Rep. 2021. PMID: 34132942 Review.
-
mTORC2: The other mTOR in autophagy regulation.Aging Cell. 2021 Aug;20(8):e13431. doi: 10.1111/acel.13431. Epub 2021 Jul 12. Aging Cell. 2021. PMID: 34250734 Free PMC article. Review.
References
-
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments With Fission Yeast: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

