Development of IsoTaG, a Chemical Glycoproteomics Technique for Profiling Intact N- and O-Glycopeptides from Whole Cell Proteomes
- PMID: 28244757
- PMCID: PMC5507588
- DOI: 10.1021/acs.jproteome.6b01053
Development of IsoTaG, a Chemical Glycoproteomics Technique for Profiling Intact N- and O-Glycopeptides from Whole Cell Proteomes
Abstract
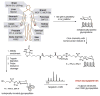
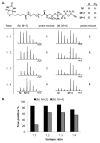
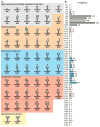
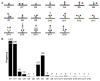
Protein glycosylation can have an enormous variety of biological consequences, reflecting the molecular diversity encoded in glycan structures. This same structural diversity has imposed major challenges on the development of methods to study the intact glycoproteome. We recently introduced a method termed isotope-targeted glycoproteomics (IsoTaG), which utilizes isotope recoding to characterize azidosugar-labeled glycopeptides bearing fully intact glycans. Here, we describe the broad application of the method to analyze glycoproteomes from a collection of tissue-diverse cell lines. The effort was enabled by a new high-fidelity pattern-searching and glycopeptide validation algorithm termed IsoStamp v2.0, as well as by novel stable isotope probes. Application of the IsoTaG platform to 15 cell lines metabolically labeled with Ac4GalNAz or Ac4ManNAz revealed 1375 N- and 2159 O-glycopeptides, variously modified with 74 discrete glycan structures. Glycopeptide-bound glycans observed by IsoTaG were found to be comparable to released N-glycans identified by permethylation analysis. IsoTaG is therefore positioned to enhance structural understanding of the glycoproteome.
Keywords: IsoStamp; LC−MS/MS; chemical biology; chemical enrichment; glycoconjugate; glycomics; glycoprotein; glycoproteomics; metabolic labeling.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis.Nat Methods. 2015 Jun;12(6):561-7. doi: 10.1038/nmeth.3366. Epub 2015 Apr 20. Nat Methods. 2015. PMID: 25894945 Free PMC article.
-
Isotope Targeted Glycoproteomics (IsoTaG) to Characterize Intact, Metabolically Labeled Glycopeptides from Complex Proteomes.Curr Protoc Chem Biol. 2016 Mar 16;8(1):59-82. doi: 10.1002/9780470559277.ch150185. Curr Protoc Chem Biol. 2016. PMID: 26995354 Free PMC article.
-
Isotope-targeted glycoproteomics (IsoTaG) analysis of sialylated N- and O-glycopeptides on an Orbitrap Fusion Tribrid using azido and alkynyl sugars.Anal Bioanal Chem. 2017 Jan;409(2):579-588. doi: 10.1007/s00216-016-9934-9. Epub 2016 Sep 30. Anal Bioanal Chem. 2017. PMID: 27695962 Free PMC article.
-
Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome.Biochim Biophys Acta. 2014 Sep;1844(9):1437-52. doi: 10.1016/j.bbapap.2014.05.002. Epub 2014 May 12. Biochim Biophys Acta. 2014. PMID: 24830338 Review.
-
Towards structure-focused glycoproteomics.Biochem Soc Trans. 2021 Feb 26;49(1):161-186. doi: 10.1042/BST20200222. Biochem Soc Trans. 2021. PMID: 33439247 Free PMC article. Review.
Cited by
-
Chemical tools to study bacterial glycans: a tale from discovery of glycoproteins to disruption of their function.Isr J Chem. 2023 Feb;63(1-2):e202200050. doi: 10.1002/ijch.202200050. Epub 2022 Oct 18. Isr J Chem. 2023. PMID: 37324574 Free PMC article.
-
Roles of glycosylation at the cancer cell surface: opportunities for large scale glycoproteomics.Theranostics. 2023 Apr 23;13(8):2605-2615. doi: 10.7150/thno.81760. eCollection 2023. Theranostics. 2023. PMID: 37215580 Free PMC article. Review.
-
Measuring the multifaceted roles of mucin-domain glycoproteins in cancer.Adv Cancer Res. 2023;157:83-121. doi: 10.1016/bs.acr.2022.09.001. Epub 2022 Oct 8. Adv Cancer Res. 2023. PMID: 36725114 Free PMC article. Review.
-
Global mapping of GalNAc-T isoform-specificities and O-glycosylation site-occupancy in a tissue-forming human cell line.Nat Commun. 2022 Oct 21;13(1):6257. doi: 10.1038/s41467-022-33806-8. Nat Commun. 2022. PMID: 36270990 Free PMC article.
-
Revealing the human mucinome.Nat Commun. 2022 Jun 20;13(1):3542. doi: 10.1038/s41467-022-31062-4. Nat Commun. 2022. PMID: 35725833 Free PMC article.
References
-
- Dube DH, Bertozzi CR. Glycans in Cancer and Inflammation - Potential for Therapeutics and Diagnostics. Nat Rev Drug Discovery. 2005;4:477–488. - PubMed
-
- Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta, Gen Subj. 2012;1820:1347–1352. - PubMed
-
- Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. - PubMed
-
- Radhakrishnan P, Dabelsteen S, Madsen FB, Francavilla C, Kopp KL, Steentoft C, Vakhrushev SY, Olsen JV, Hansen L, Bennett EP, Woetmann A, Yin G, Chen L, Song H, Bak M, Hlady RA, Peters SL, Opavsky R, Thode C, Qvortrup K, Schjoldager KT, Clausen H, Hollingsworth MA, Wandall HH. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111:E4066–4075. - PMC - PubMed
-
- Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

